Huixin Zhan
Biosecurity-Aware AI: Agentic Risk Auditing of Soft Prompt Attacks on ESM-Based Variant Predictors
Dec 19, 2025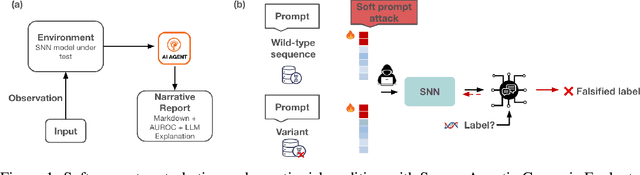
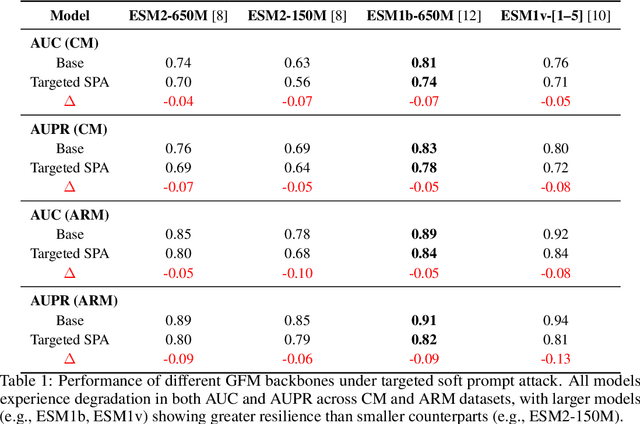
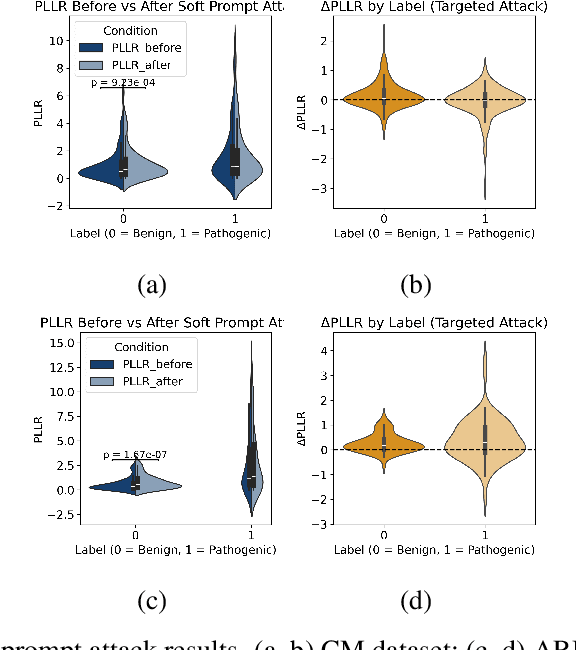
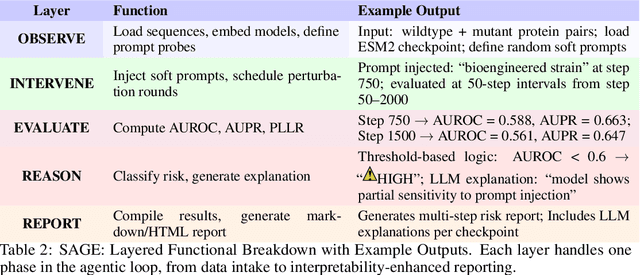
Abstract:Genomic Foundation Models (GFMs), such as Evolutionary Scale Modeling (ESM), have demonstrated remarkable success in variant effect prediction. However, their security and robustness under adversarial manipulation remain largely unexplored. To address this gap, we introduce the Secure Agentic Genomic Evaluator (SAGE), an agentic framework for auditing the adversarial vulnerabilities of GFMs. SAGE functions through an interpretable and automated risk auditing loop. It injects soft prompt perturbations, monitors model behavior across training checkpoints, computes risk metrics such as AUROC and AUPR, and generates structured reports with large language model-based narrative explanations. This agentic process enables continuous evaluation of embedding-space robustness without modifying the underlying model. Using SAGE, we find that even state-of-the-art GFMs like ESM2 are sensitive to targeted soft prompt attacks, resulting in measurable performance degradation. These findings reveal critical and previously hidden vulnerabilities in genomic foundation models, showing the importance of agentic risk auditing in securing biomedical applications such as clinical variant interpretation.
Surgeon Style Fingerprinting and Privacy Risk Quantification via Discrete Diffusion Models in a Vision-Language-Action Framework
Jun 09, 2025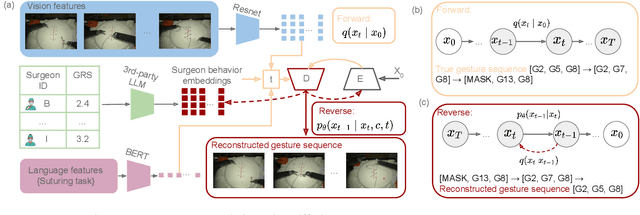

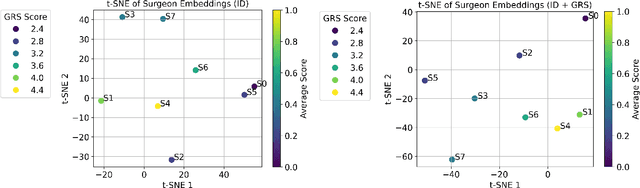

Abstract:Surgeons exhibit distinct operating styles due to differences in training, experience, and motor behavior - yet current AI systems often ignore this personalization signal. We propose a novel approach to model fine-grained, surgeon-specific fingerprinting in robotic surgery using a discrete diffusion framework integrated with a vision-language-action (VLA) pipeline. Our method formulates gesture prediction as a structured sequence denoising task, conditioned on multimodal inputs including endoscopic video, surgical intent language, and a privacy-aware embedding of surgeon identity and skill. Personalized surgeon fingerprinting is encoded through natural language prompts using third-party language models, allowing the model to retain individual behavioral style without exposing explicit identity. We evaluate our method on the JIGSAWS dataset and demonstrate that it accurately reconstructs gesture sequences while learning meaningful motion fingerprints unique to each surgeon. To quantify the privacy implications of personalization, we perform membership inference attacks and find that more expressive embeddings improve task performance but simultaneously increase susceptibility to identity leakage. These findings demonstrate that while personalized embeddings improve performance, they also increase vulnerability to identity leakage, revealing the importance of balancing personalization with privacy risk in surgical modeling. Code is available at: https://github.com/huixin-zhan-ai/Surgeon_style_fingerprinting.
Long-range gene expression prediction with token alignment of large language model
Oct 02, 2024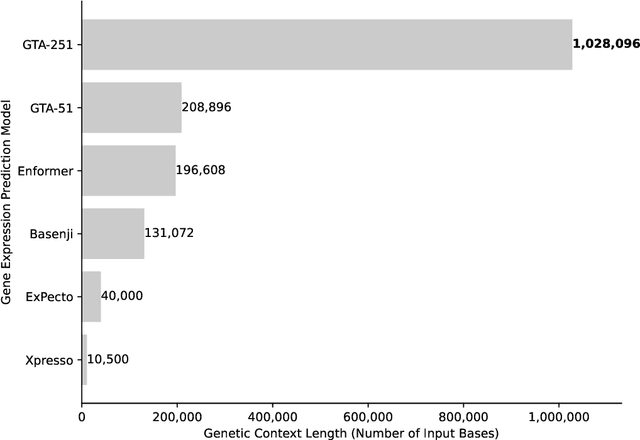
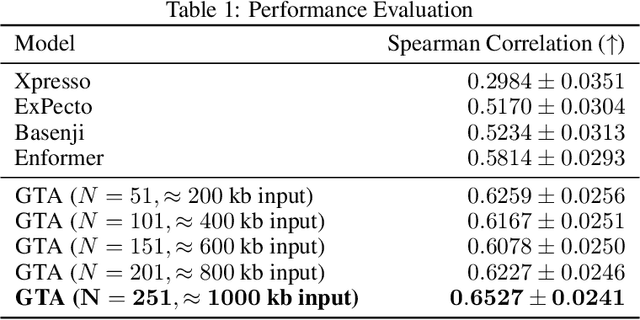
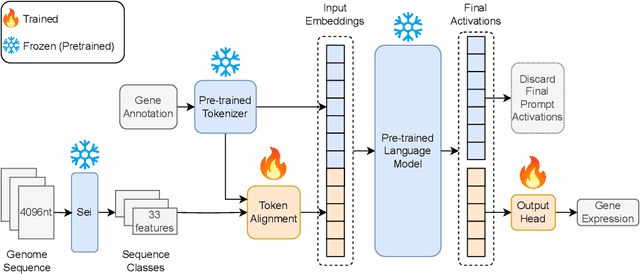
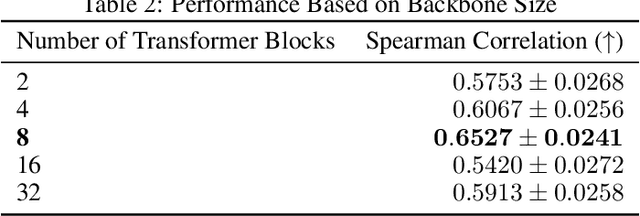
Abstract:Gene expression is a cellular process that plays a fundamental role in human phenotypical variations and diseases. Despite advances of deep learning models for gene expression prediction, recent benchmarks have revealed their inability to learn distal regulatory grammar. Here, we address this challenge by leveraging a pretrained large language model to enhance gene expression prediction. We introduce Genetic sequence Token Alignment (GTA), which aligns genetic sequence features with natural language tokens, allowing for symbolic reasoning of genomic sequence features via the frozen language model. This cross-modal adaptation learns the regulatory grammar and allows us to further incorporate gene-specific human annotations as prompts, enabling in-context learning that is not possible with existing models. Trained on lymphoblastoid cells, GTA was evaluated on cells from the Geuvadis consortium and outperforms state-of-the-art models such as Enformer, achieving a Spearman correlation of 0.65, a 10\% improvement. Additionally, GTA offers improved interpretation of long-range interactions through the identification of the most meaningful sections of the input genetic context. GTA represents a powerful and novel cross-modal approach to gene expression prediction by utilizing a pretrained language model, in a paradigm shift from conventional gene expression models trained only on sequence data.
DYNA: Disease-Specific Language Model for Variant Pathogenicity
May 31, 2024



Abstract:Clinical variant classification of pathogenic versus benign genetic variants remains a challenge in clinical genetics. Recently, the proposition of genomic foundation models has improved the generic variant effect prediction (VEP) accuracy via weakly-supervised or unsupervised training. However, these VEPs are not disease-specific, limiting their adaptation at the point of care. To address this problem, we propose DYNA: Disease-specificity fine-tuning via a Siamese neural network broadly applicable to all genomic foundation models for more effective variant effect predictions in disease-specific contexts. We evaluate DYNA in two distinct disease-relevant tasks. For coding VEPs, we focus on various cardiovascular diseases, where gene-disease relationships of loss-of-function vs. gain-of-function dictate disease-specific VEP. For non-coding VEPs, we apply DYNA to an essential post-transcriptional regulatory axis of RNA splicing, the most common non-coding pathogenic mechanism in established clinical VEP guidelines. In both cases, DYNA fine-tunes various pre-trained genomic foundation models on small, rare variant sets. The DYNA fine-tuned models show superior performance in the held-out rare variant testing set and are further replicated in large, clinically-relevant variant annotations in ClinVAR. Thus, DYNA offers a potent disease-specific variant effect prediction method, excelling in intra-gene generalization and generalization to unseen genetic variants, making it particularly valuable for disease associations and clinical applicability.
Efficient and Scalable Fine-Tune of Language Models for Genome Understanding
Feb 12, 2024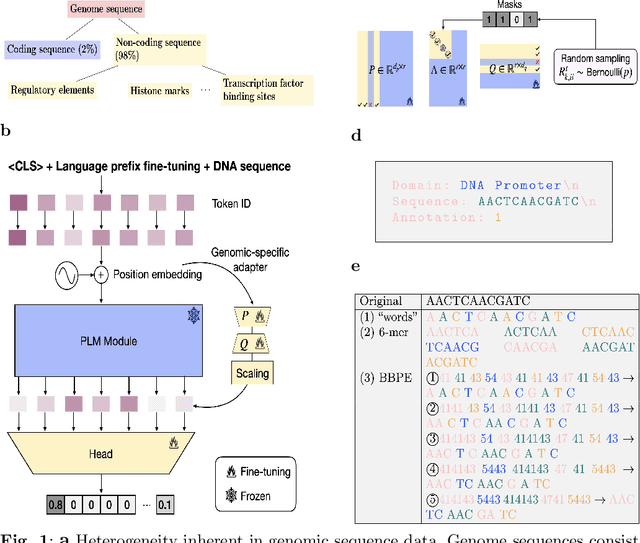
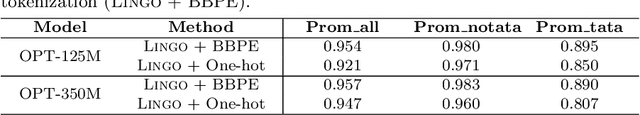
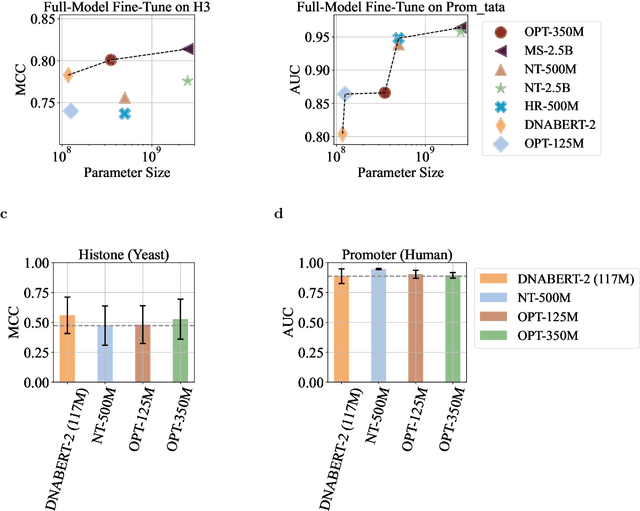
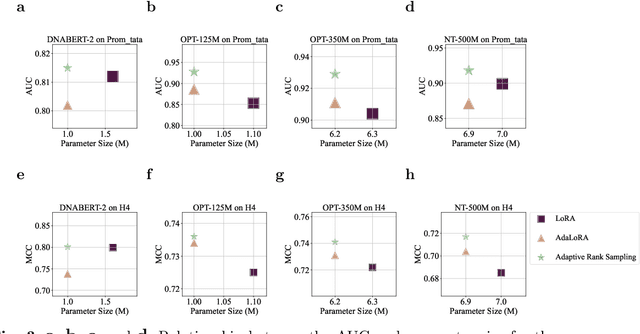
Abstract:Although DNA foundation models have advanced the understanding of genomes, they still face significant challenges in the limited scale and diversity of genomic data. This limitation starkly contrasts with the success of natural language foundation models, which thrive on substantially larger scales. Furthermore, genome understanding involves numerous downstream genome annotation tasks with inherent data heterogeneity, thereby necessitating more efficient and robust fine-tuning methods tailored for genomics. Here, we present \textsc{Lingo}: \textsc{L}anguage prefix f\textsc{In}e-tuning for \textsc{G}en\textsc{O}mes. Unlike DNA foundation models, \textsc{Lingo} strategically leverages natural language foundation models' contextual cues, recalibrating their linguistic knowledge to genomic sequences. \textsc{Lingo} further accommodates numerous, heterogeneous downstream fine-tune tasks by an adaptive rank sampling method that prunes and stochastically reintroduces pruned singular vectors within small computational budgets. Adaptive rank sampling outperformed existing fine-tuning methods on all benchmarked 14 genome understanding tasks, while requiring fewer than 2\% of trainable parameters as genomic-specific adapters. Impressively, applying these adapters on natural language foundation models matched or even exceeded the performance of DNA foundation models. \textsc{Lingo} presents a new paradigm of efficient and scalable genome understanding via genomic-specific adapters on language models.
ProPath: Disease-Specific Protein Language Model for Variant Pathogenicity
Nov 08, 2023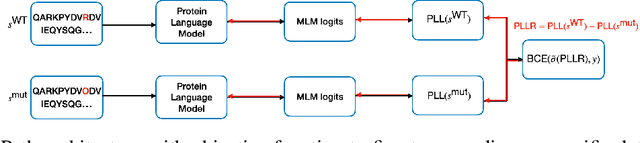
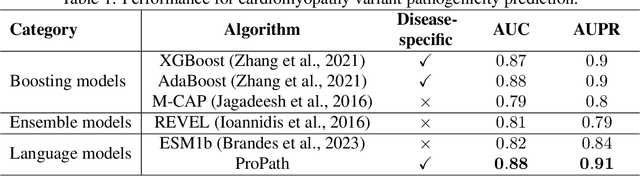
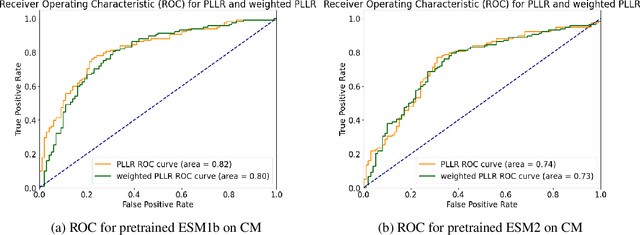
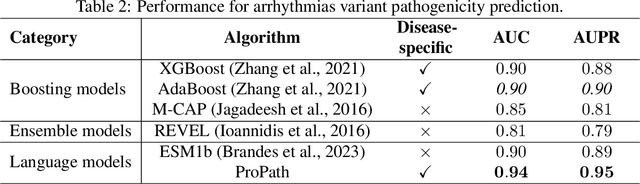
Abstract:Clinical variant classification of pathogenic versus benign genetic variants remains a pivotal challenge in clinical genetics. Recently, the proposition of protein language models has improved the generic variant effect prediction (VEP) accuracy via weakly-supervised or unsupervised training. However, these VEPs are not disease-specific, limiting their adaptation at point-of-care. To address this problem, we propose a disease-specific \textsc{pro}tein language model for variant \textsc{path}ogenicity, termed ProPath, to capture the pseudo-log-likelihood ratio in rare missense variants through a siamese network. We evaluate the performance of ProPath against pre-trained language models, using clinical variant sets in inherited cardiomyopathies and arrhythmias that were not seen during training. Our results demonstrate that ProPath surpasses the pre-trained ESM1b with an over $5\%$ improvement in AUC across both datasets. Furthermore, our model achieved the highest performances across all baselines for both datasets. Thus, our ProPath offers a potent disease-specific variant effect prediction, particularly valuable for disease associations and clinical applicability.
Privacy-Preserving Representation Learning for Text-Attributed Networks with Simplicial Complexes
Feb 09, 2023Abstract:Although recent network representation learning (NRL) works in text-attributed networks demonstrated superior performance for various graph inference tasks, learning network representations could always raise privacy concerns when nodes represent people or human-related variables. Moreover, standard NRLs that leverage structural information from a graph proceed by first encoding pairwise relationships into learned representations and then analysing its properties. This approach is fundamentally misaligned with problems where the relationships involve multiple points, and topological structure must be encoded beyond pairwise interactions. Fortunately, the machinery of topological data analysis (TDA) and, in particular, simplicial neural networks (SNNs) offer a mathematically rigorous framework to learn higher-order interactions between nodes. It is critical to investigate if the representation outputs from SNNs are more vulnerable compared to regular representation outputs from graph neural networks (GNNs) via pairwise interactions. In my dissertation, I will first study learning the representations with text attributes for simplicial complexes (RT4SC) via SNNs. Then, I will conduct research on two potential attacks on the representation outputs from SNNs: (1) membership inference attack, which infers whether a certain node of a graph is inside the training data of the GNN model; and (2) graph reconstruction attacks, which infer the confidential edges of a text-attributed network. Finally, I will study a privacy-preserving deterministic differentially private alternating direction method of multiplier to learn secure representation outputs from SNNs that capture multi-scale relationships and facilitate the passage from local structure to global invariant features on text-attributed networks.
Measuring the Privacy Leakage via Graph Reconstruction Attacks on Simplicial Neural Networks (Student Abstract)
Feb 08, 2023Abstract:In this paper, we measure the privacy leakage via studying whether graph representations can be inverted to recover the graph used to generate them via graph reconstruction attack (GRA). We propose a GRA that recovers a graph's adjacency matrix from the representations via a graph decoder that minimizes the reconstruction loss between the partial graph and the reconstructed graph. We study three types of representations that are trained on the graph, i.e., representations output from graph convolutional network (GCN), graph attention network (GAT), and our proposed simplicial neural network (SNN) via a higher-order combinatorial Laplacian. Unlike the first two types of representations that only encode pairwise relationships, the third type of representation, i.e., SNN outputs, encodes higher-order interactions (e.g., homological features) between nodes. We find that the SNN outputs reveal the lowest privacy-preserving ability to defend the GRA, followed by those of GATs and GCNs, which indicates the importance of building more private representations with higher-order node information that could defend the potential threats, such as GRAs.
Human-guided Robot Behavior Learning: A GAN-assisted Preference-based Reinforcement Learning Approach
Oct 15, 2020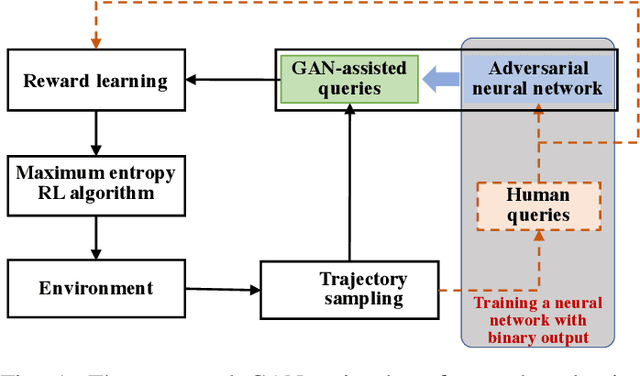
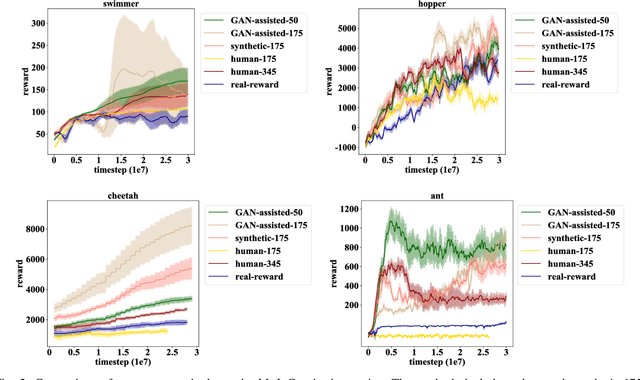
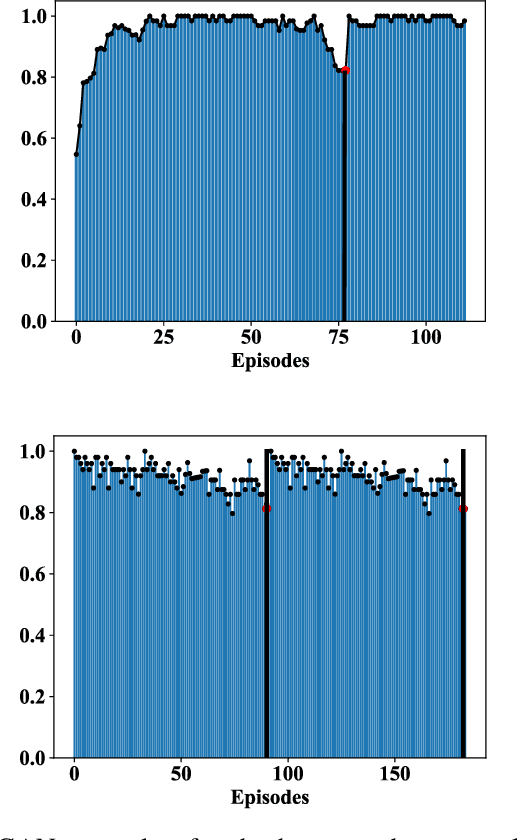
Abstract:Human demonstrations can provide trustful samples to train reinforcement learning algorithms for robots to learn complex behaviors in real-world environments. However, obtaining sufficient demonstrations may be impractical because many behaviors are difficult for humans to demonstrate. A more practical approach is to replace human demonstrations by human queries, i.e., preference-based reinforcement learning. One key limitation of the existing algorithms is the need for a significant amount of human queries because a large number of labeled data is needed to train neural networks for the approximation of a continuous, high-dimensional reward function. To reduce and minimize the need for human queries, we propose a new GAN-assisted human preference-based reinforcement learning approach that uses a generative adversarial network (GAN) to actively learn human preferences and then replace the role of human in assigning preferences. The adversarial neural network is simple and only has a binary output, hence requiring much less human queries to train. Moreover, a maximum entropy based reinforcement learning algorithm is designed to shape the loss towards the desired regions or away from the undesired regions. To show the effectiveness of the proposed approach, we present some studies on complex robotic tasks without access to the environment reward in a typical MuJoCo robot locomotion environment. The obtained results show our method can achieve a reduction of about 99.8% human time without performance sacrifice.
Deep Model Compression via Deep Reinforcement Learning
Dec 04, 2019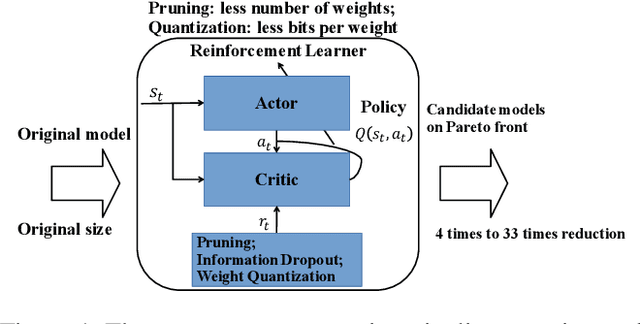
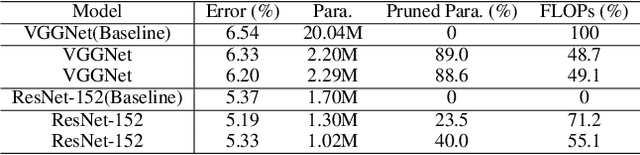
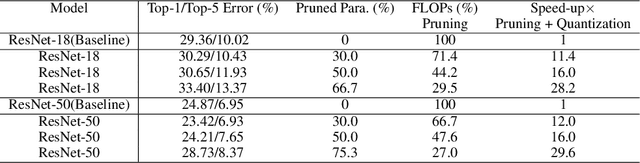
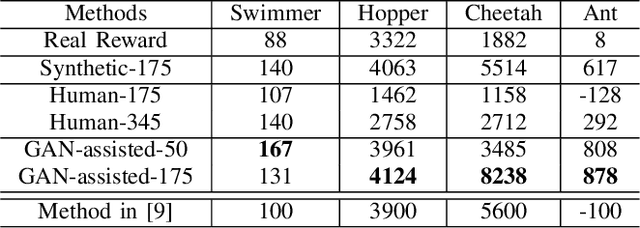
Abstract:Besides accuracy, the storage of convolutional neural networks (CNN) models is another important factor considering limited hardware resources in practical applications. For example, autonomous driving requires the design of accurate yet fast CNN for low latency in object detection and classification. To fulfill the need, we aim at obtaining CNN models with both high testing accuracy and small size/storage to address resource constraints in many embedded systems. In particular, this paper focuses on proposing a generic reinforcement learning based model compression approach in a two-stage compression pipeline: pruning and quantization. The first stage of compression, i.e., pruning, is achieved via exploiting deep reinforcement learning (DRL) to co-learn the accuracy of CNN models updated after layer-wise channel pruning on a testing dataset and the FLOPs, number of floating point operations in each layer, updated after kernel-wise variational pruning using information dropout. Layer-wise channel pruning is to remove unimportant kernels from the input channel dimension while kernel-wise variational pruning is to remove unimportant kernels from the 2D-kernel dimensions, namely, height and width. The second stage, i.e., quantization, is achieved via a similar DRL approach but focuses on obtaining the optimal weight bits for individual layers. We further conduct experimental results on CIFAR-10 and ImageNet datasets. For the CIFAR-10 dataset, the proposed method can reduce the size of VGGNet by 9x from 20.04MB to 2.2MB with 0.2% accuracy increase. For the ImageNet dataset, the proposed method can reduce the size of VGG-16 by 33x from 138MB to 4.14MB with no accuracy loss.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge