Danyi Li
Enhancing WSI-Based Survival Analysis with Report-Auxiliary Self-Distillation
Sep 19, 2025Abstract:Survival analysis based on Whole Slide Images (WSIs) is crucial for evaluating cancer prognosis, as they offer detailed microscopic information essential for predicting patient outcomes. However, traditional WSI-based survival analysis usually faces noisy features and limited data accessibility, hindering their ability to capture critical prognostic features effectively. Although pathology reports provide rich patient-specific information that could assist analysis, their potential to enhance WSI-based survival analysis remains largely unexplored. To this end, this paper proposes a novel Report-auxiliary self-distillation (Rasa) framework for WSI-based survival analysis. First, advanced large language models (LLMs) are utilized to extract fine-grained, WSI-relevant textual descriptions from original noisy pathology reports via a carefully designed task prompt. Next, a self-distillation-based pipeline is designed to filter out irrelevant or redundant WSI features for the student model under the guidance of the teacher model's textual knowledge. Finally, a risk-aware mix-up strategy is incorporated during the training of the student model to enhance both the quantity and diversity of the training data. Extensive experiments carried out on our collected data (CRC) and public data (TCGA-BRCA) demonstrate the superior effectiveness of Rasa against state-of-the-art methods. Our code is available at https://github.com/zhengwang9/Rasa.
Few-Shot Learning for Annotation-Efficient Nucleus Instance Segmentation
Feb 28, 2024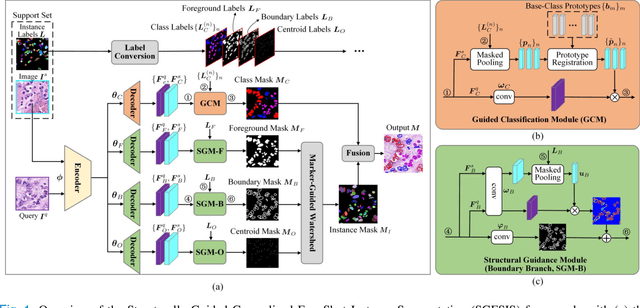
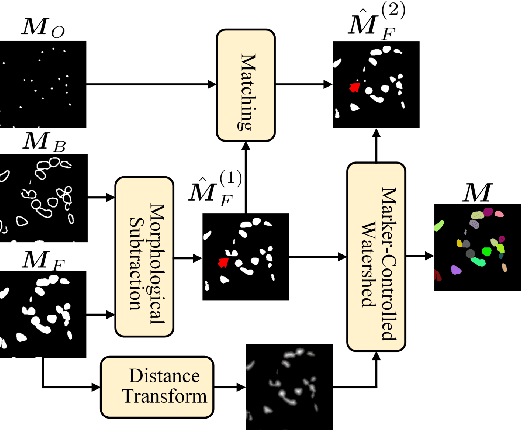
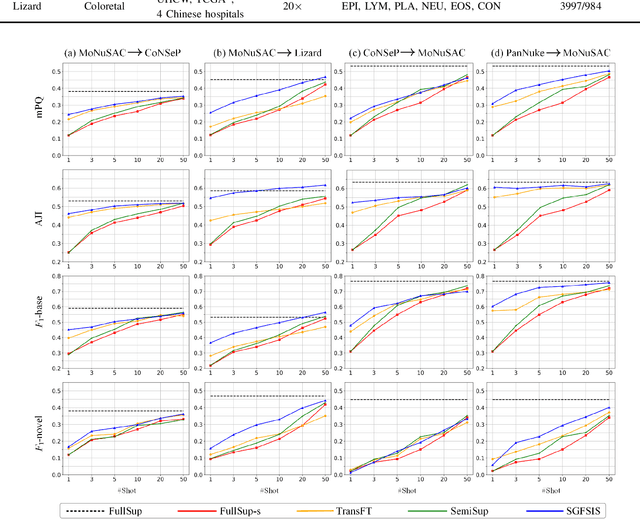
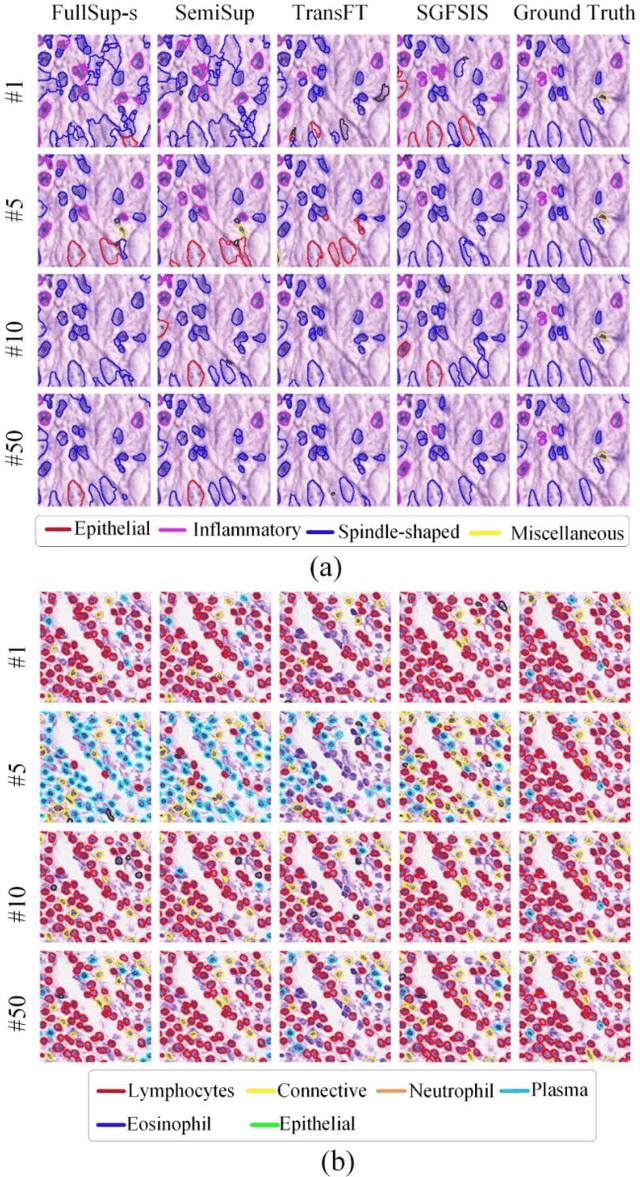
Abstract:Nucleus instance segmentation from histopathology images suffers from the extremely laborious and expert-dependent annotation of nucleus instances. As a promising solution to this task, annotation-efficient deep learning paradigms have recently attracted much research interest, such as weakly-/semi-supervised learning, generative adversarial learning, etc. In this paper, we propose to formulate annotation-efficient nucleus instance segmentation from the perspective of few-shot learning (FSL). Our work was motivated by that, with the prosperity of computational pathology, an increasing number of fully-annotated datasets are publicly accessible, and we hope to leverage these external datasets to assist nucleus instance segmentation on the target dataset which only has very limited annotation. To achieve this goal, we adopt the meta-learning based FSL paradigm, which however has to be tailored in two substantial aspects before adapting to our task. First, since the novel classes may be inconsistent with those of the external dataset, we extend the basic definition of few-shot instance segmentation (FSIS) to generalized few-shot instance segmentation (GFSIS). Second, to cope with the intrinsic challenges of nucleus segmentation, including touching between adjacent cells, cellular heterogeneity, etc., we further introduce a structural guidance mechanism into the GFSIS network, finally leading to a unified Structurally-Guided Generalized Few-Shot Instance Segmentation (SGFSIS) framework. Extensive experiments on a couple of publicly accessible datasets demonstrate that, SGFSIS can outperform other annotation-efficient learning baselines, including semi-supervised learning, simple transfer learning, etc., with comparable performance to fully supervised learning with less than 5% annotations.
Rethinking Mitosis Detection: Towards Diverse Data and Feature Representation
Jul 12, 2023Abstract:Mitosis detection is one of the fundamental tasks in computational pathology, which is extremely challenging due to the heterogeneity of mitotic cell. Most of the current studies solve the heterogeneity in the technical aspect by increasing the model complexity. However, lacking consideration of the biological knowledge and the complex model design may lead to the overfitting problem while limited the generalizability of the detection model. In this paper, we systematically study the morphological appearances in different mitotic phases as well as the ambiguous non-mitotic cells and identify that balancing the data and feature diversity can achieve better generalizability. Based on this observation, we propose a novel generalizable framework (MitDet) for mitosis detection. The data diversity is considered by the proposed diversity-guided sample balancing (DGSB). And the feature diversity is preserved by inter- and intra- class feature diversity-preserved module (InCDP). Stain enhancement (SE) module is introduced to enhance the domain-relevant diversity of both data and features simultaneously. Extensive experiments have demonstrated that our proposed model outperforms all the SOTA approaches in several popular mitosis detection datasets in both internal and external test sets using minimal annotation efforts with point annotations only. Comprehensive ablation studies have also proven the effectiveness of the rethinking of data and feature diversity balancing. By analyzing the results quantitatively and qualitatively, we believe that our proposed model not only achieves SOTA performance but also might inspire the future studies in new perspectives. Source code is at https://github.com/Onehour0108/MitDet.
RestainNet: a self-supervised digital re-stainer for stain normalization
Feb 28, 2022



Abstract:Color inconsistency is an inevitable challenge in computational pathology, which generally happens because of stain intensity variations or sections scanned by different scanners. It harms the pathological image analysis methods, especially the learning-based models. A series of approaches have been proposed for stain normalization. However, most of them are lack flexibility in practice. In this paper, we formulated stain normalization as a digital re-staining process and proposed a self-supervised learning model, which is called RestainNet. Our network is regarded as a digital restainer which learns how to re-stain an unstained (grayscale) image. Two digital stains, Hematoxylin (H) and Eosin (E) were extracted from the original image by Beer-Lambert's Law. We proposed a staining loss to maintain the correctness of stain intensity during the restaining process. Thanks to the self-supervised nature, paired training samples are no longer necessary, which demonstrates great flexibility in practical usage. Our RestainNet outperforms existing approaches and achieves state-of-the-art performance with regard to color correctness and structure preservation. We further conducted experiments on the segmentation and classification tasks and the proposed RestainNet achieved outstanding performance compared with SOTA methods. The self-supervised design allows the network to learn any staining style with no extra effort.
PDBL: Improving Histopathological Tissue Classification with Plug-and-Play Pyramidal Deep-Broad Learning
Nov 04, 2021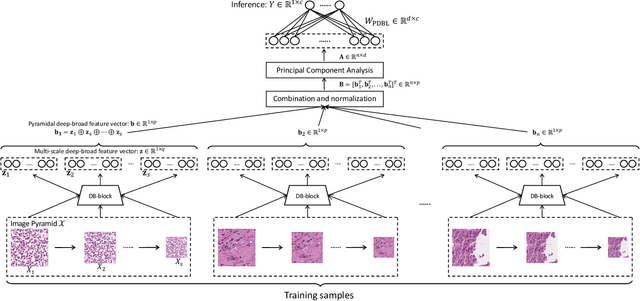
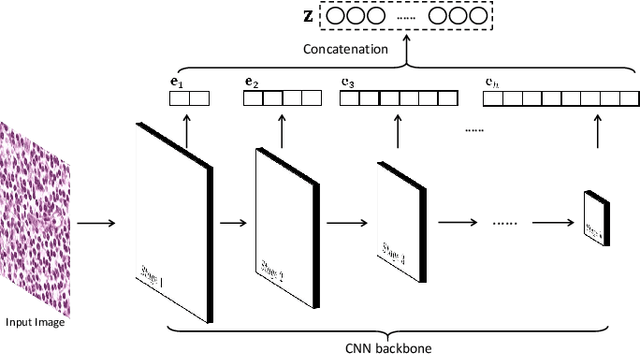
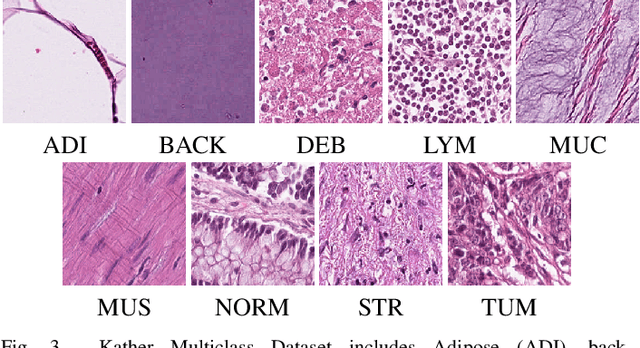
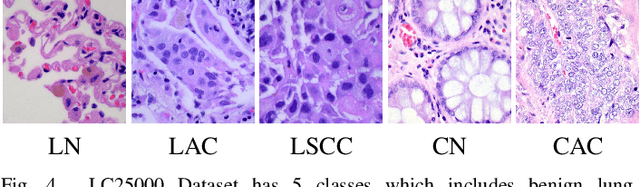
Abstract:Histopathological tissue classification is a fundamental task in pathomics cancer research. Precisely differentiating different tissue types is a benefit for the downstream researches, like cancer diagnosis, prognosis and etc. Existing works mostly leverage the popular classification backbones in computer vision to achieve histopathological tissue classification. In this paper, we proposed a super lightweight plug-and-play module, named Pyramidal Deep-Broad Learning (PDBL), for any well-trained classification backbone to further improve the classification performance without a re-training burden. We mimic how pathologists observe pathology slides in different magnifications and construct an image pyramid for the input image in order to obtain the pyramidal contextual information. For each level in the pyramid, we extract the multi-scale deep-broad features by our proposed Deep-Broad block (DB-block). We equipped PDBL in three popular classification backbones, ShuffLeNetV2, EfficientNetb0, and ResNet50 to evaluate the effectiveness and efficiency of our proposed module on two datasets (Kather Multiclass Dataset and the LC25000 Dataset). Experimental results demonstrate the proposed PDBL can steadily improve the tissue-level classification performance for any CNN backbones, especially for the lightweight models when given a small among of training samples (less than 10%), which greatly saves the computational time and annotation efforts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge