Dali Wang
ORBIT-2: Scaling Exascale Vision Foundation Models for Weather and Climate Downscaling
May 07, 2025



Abstract:Sparse observations and coarse-resolution climate models limit effective regional decision-making, underscoring the need for robust downscaling. However, existing AI methods struggle with generalization across variables and geographies and are constrained by the quadratic complexity of Vision Transformer (ViT) self-attention. We introduce ORBIT-2, a scalable foundation model for global, hyper-resolution climate downscaling. ORBIT-2 incorporates two key innovations: (1) Residual Slim ViT (Reslim), a lightweight architecture with residual learning and Bayesian regularization for efficient, robust prediction; and (2) TILES, a tile-wise sequence scaling algorithm that reduces self-attention complexity from quadratic to linear, enabling long-sequence processing and massive parallelism. ORBIT-2 scales to 10 billion parameters across 32,768 GPUs, achieving up to 1.8 ExaFLOPS sustained throughput and 92-98% strong scaling efficiency. It supports downscaling to 0.9 km global resolution and processes sequences up to 4.2 billion tokens. On 7 km resolution benchmarks, ORBIT-2 achieves high accuracy with R^2 scores in the range of 0.98 to 0.99 against observation data.
S3LLM: Large-Scale Scientific Software Understanding with LLMs using Source, Metadata, and Document
Mar 15, 2024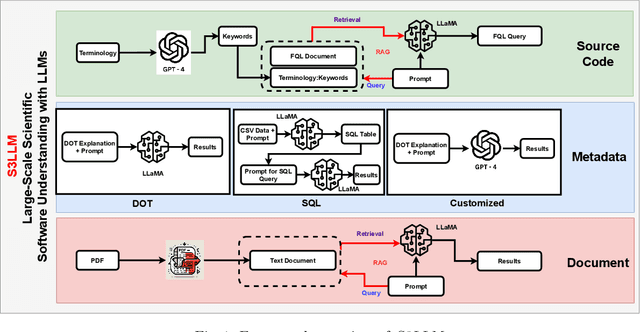
Abstract:The understanding of large-scale scientific software poses significant challenges due to its diverse codebase, extensive code length, and target computing architectures. The emergence of generative AI, specifically large language models (LLMs), provides novel pathways for understanding such complex scientific codes. This paper presents S3LLM, an LLM-based framework designed to enable the examination of source code, code metadata, and summarized information in conjunction with textual technical reports in an interactive, conversational manner through a user-friendly interface. S3LLM leverages open-source LLaMA-2 models to enhance code analysis through the automatic transformation of natural language queries into domain-specific language (DSL) queries. Specifically, it translates these queries into Feature Query Language (FQL), enabling efficient scanning and parsing of entire code repositories. In addition, S3LLM is equipped to handle diverse metadata types, including DOT, SQL, and customized formats. Furthermore, S3LLM incorporates retrieval augmented generation (RAG) and LangChain technologies to directly query extensive documents. S3LLM demonstrates the potential of using locally deployed open-source LLMs for the rapid understanding of large-scale scientific computing software, eliminating the need for extensive coding expertise, and thereby making the process more efficient and effective. S3LLM is available at https://github.com/ResponsibleAILab/s3llm.
Geometrically Matched Multi-source Microscopic Image Synthesis Using Bidirectional Adversarial Networks
Oct 26, 2020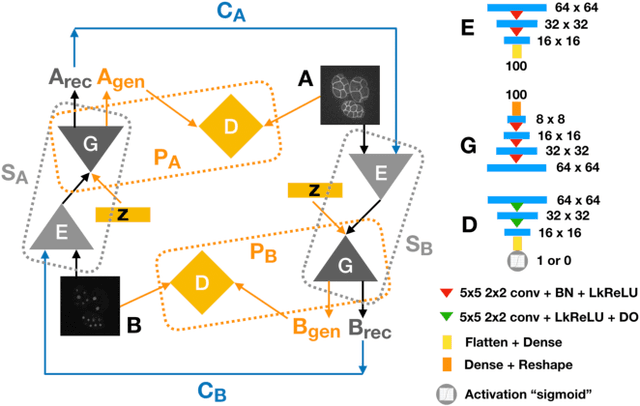

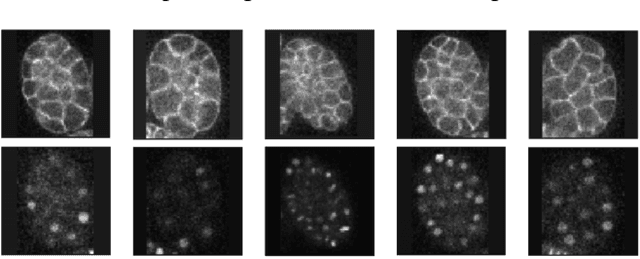
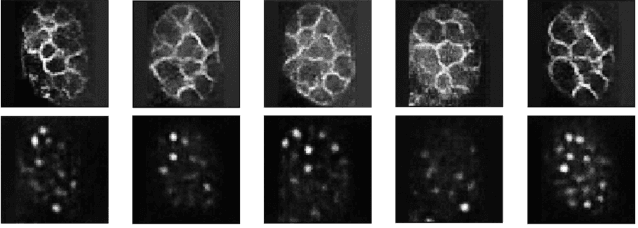
Abstract:Microscopic images from different modality can provide more complete experimental information. In practice, biological and physical limitations may prohibit the acquisition of enough microscopic images at a given observation period. Image synthesis is one promising solution. However, most existing data synthesis methods only translate the image from a source domain to a target domain without strong geometric correlations. To address this issue, we propose a novel model to synthesize diversified microscopic images from multi-sources with different geometric features. The application of our model to a 3D live time-lapse embryonic images of C. elegans presents favorable results. To the best of our knowledge, it is the first effort to synthesize microscopic images with strong underlie geometric correlations from multi-source domains that of entirely separated spatial features.
MineNav: An Expandable Synthetic Dataset Based on Minecraft for Aircraft Visual Navigation
Aug 19, 2020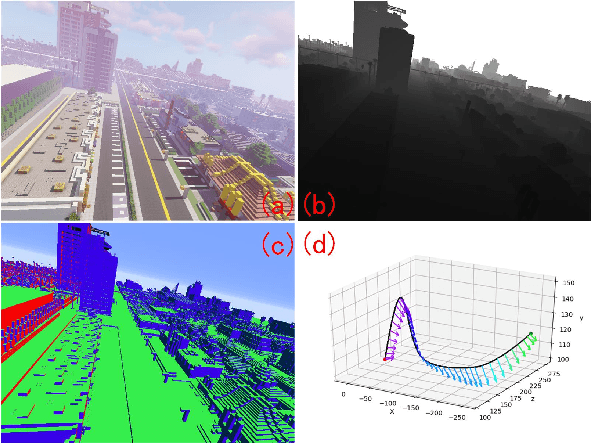
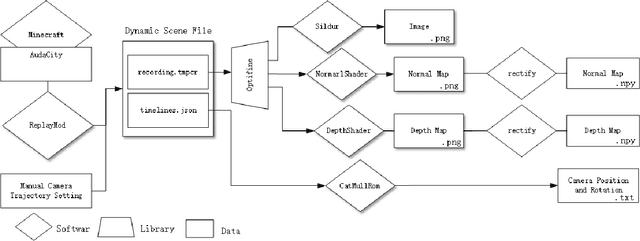
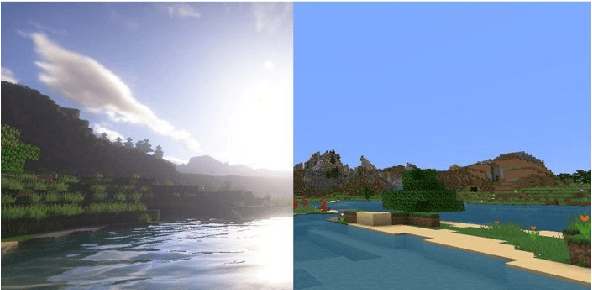
Abstract:We propose a simply method to generate high quality synthetic dataset based on open-source game Minecraft includes rendered image, Depth map, surface normal map, and 6-dof camera trajectory. This dataset has a perfect ground-truth generated by plug-in program, and thanks for the large game's community, there is an extremely large number of 3D open-world environment, users can find suitable scenes for shooting and build data sets through it and they can also build scenes in-game. as such, We don't need to worry about manual over fitting caused by too small datasets. what's more, there is also a shader community which We can use to minimize data bias between rendered images and real-images as little as possible. Last but not least, we now provide three tools to generate the data for depth prediction ,surface normal prediction and visual odometry, user can also develop the plug-in module for other vision task like segmentation or optical flow prediction.
Augmenting C. elegans Microscopic Dataset for Accelerated Pattern Recognition
May 31, 2019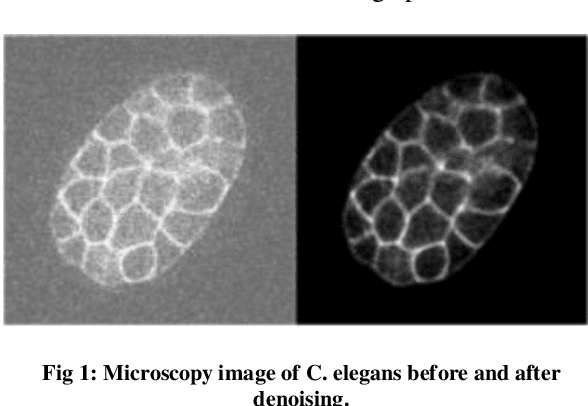
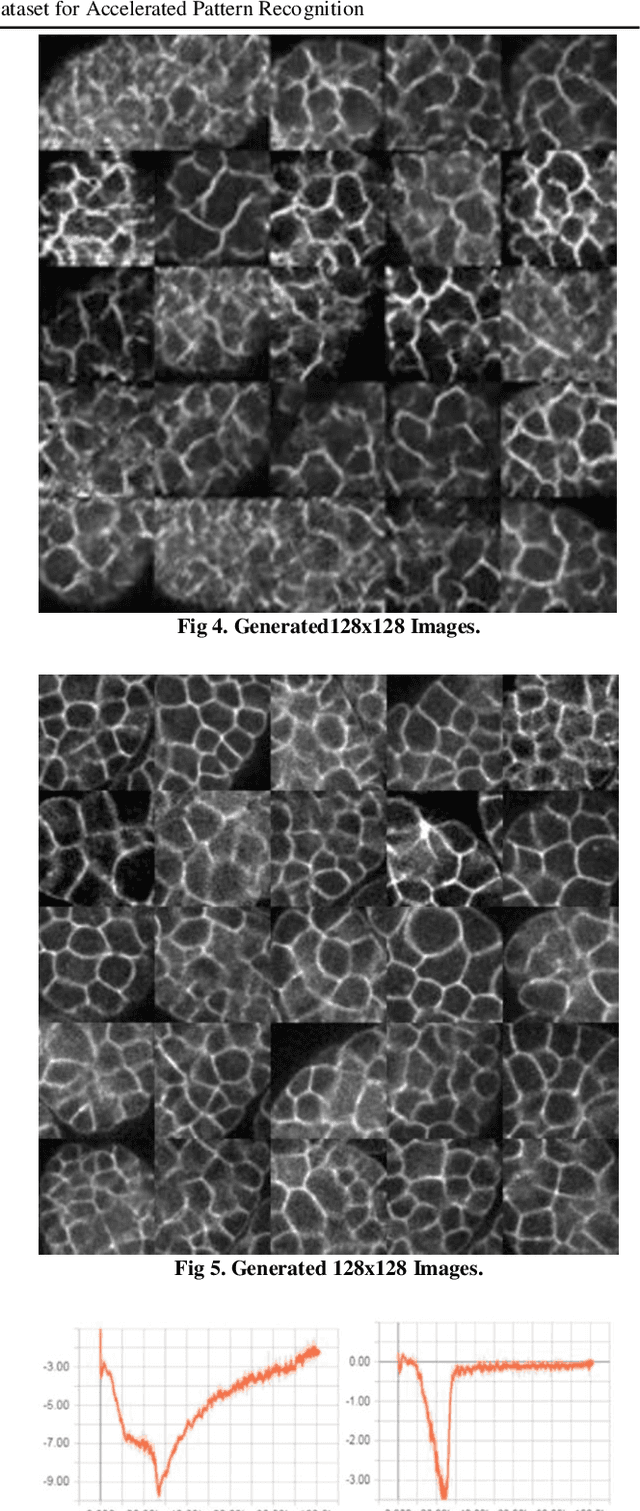
Abstract:The detection of cell shape changes in 3D time-lapse images of complex tissues is an important task. However, it is a challenging and tedious task to establish a comprehensive dataset to improve the performance of deep learning models. In the paper, we present a deep learning approach to augment 3D live images of the Caenorhabditis elegans embryo, so that we can further speed up the specific structural pattern recognition. We use an unsupervised training over unlabeled images to generate supplementary datasets for further pattern recognition. Technically, we used Alex-style neural networks in a generative adversarial network framework to generate new datasets that have common features of the C. elegans membrane structure. We also made the dataset available for a broad scientific community.
Investigating Channel Pruning through Structural Redundancy Reduction - A Statistical Study
May 19, 2019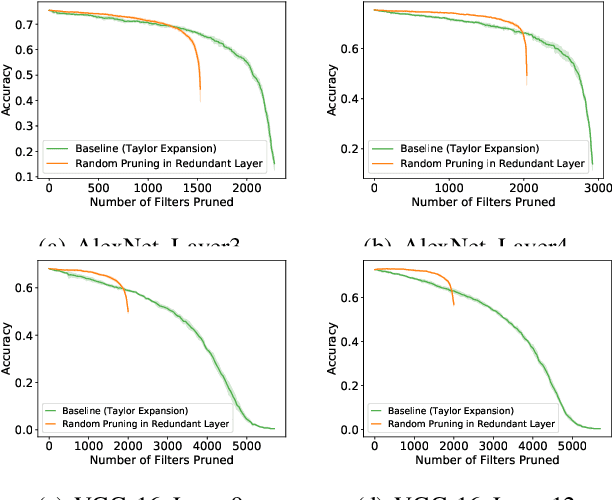
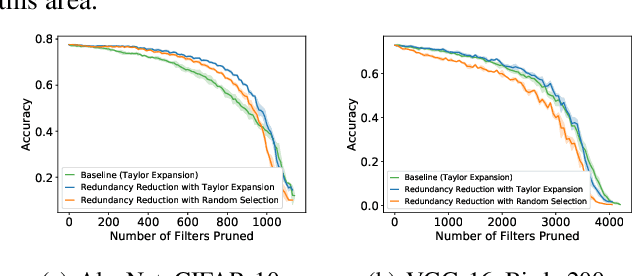
Abstract:Most existing channel pruning methods formulate the pruning task from a perspective of inefficiency reduction which iteratively rank and remove the least important filters, or find the set of filters that minimizes some reconstruction errors after pruning. In this work, we investigate the channel pruning from a new perspective with statistical modeling. We hypothesize that the number of filters at a certain layer reflects the level of 'redundancy' in that layer and thus formulate the pruning problem from the aspect of redundancy reduction. Based on both theoretic analysis and empirical studies, we make an important discovery: randomly pruning filters from layers of high redundancy outperforms pruning the least important filters across all layers based on the state-of-the-art ranking criterion. These results advance our understanding of pruning and further testify to the recent findings that the structure of the pruned model plays a key role in the network efficiency as compared to inherited weights.
Speeding up convolutional networks pruning with coarse ranking
Feb 18, 2019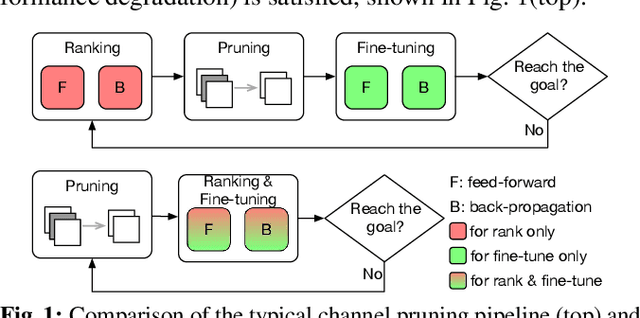
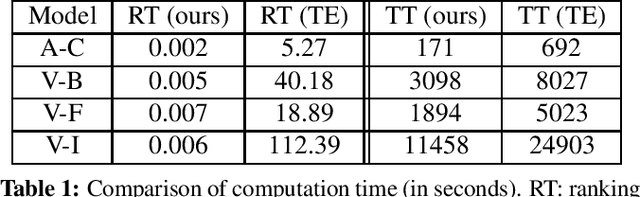
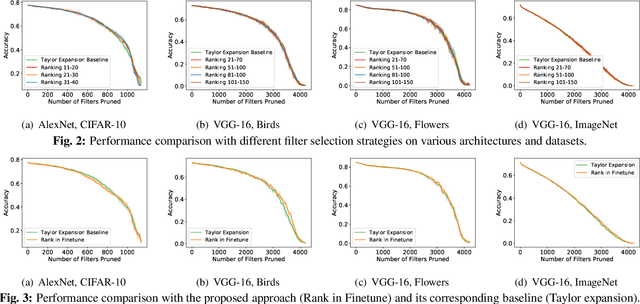
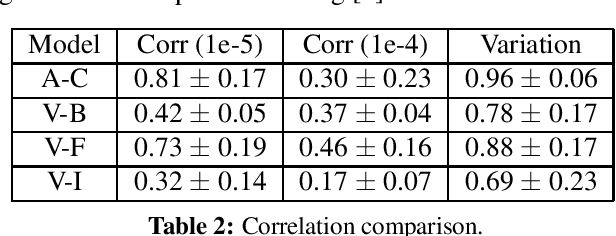
Abstract:Channel-based pruning has achieved significant successes in accelerating deep convolutional neural network, whose pipeline is an iterative three-step procedure: ranking, pruning and fine-tuning. However, this iterative procedure is computationally expensive. In this study, we present a novel computationally efficient channel pruning approach based on the coarse ranking that utilizes the intermediate results during fine-tuning to rank the importance of filters, built upon state-of-the-art works with data-driven ranking criteria. The goal of this work is not to propose a single improved approach built upon a specific channel pruning method, but to introduce a new general framework that works for a series of channel pruning methods. Various benchmark image datasets (CIFAR-10, ImageNet, Birds-200, and Flowers-102) and network architectures (AlexNet and VGG-16) are utilized to evaluate the proposed approach for object classification purpose. Experimental results show that the proposed method can achieve almost identical performance with the corresponding state-of-the-art works (baseline) while our ranking time is negligibly short. In specific, with the proposed method, 75% and 54% of the total computation time for the whole pruning procedure can be reduced for AlexNet on CIFAR-10, and for VGG-16 on ImageNet, respectively. Our approach would significantly facilitate pruning practice, especially on resource-constrained platforms.
Deep Reinforcement Learning of Cell Movement in the Early Stage of C. elegans Embryogenesis
Mar 02, 2018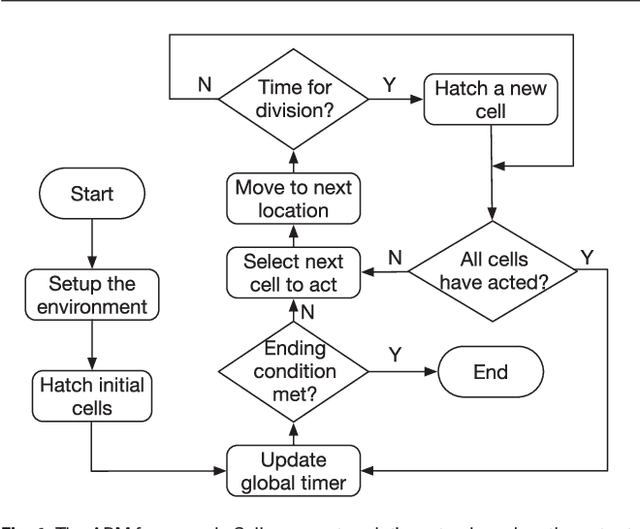
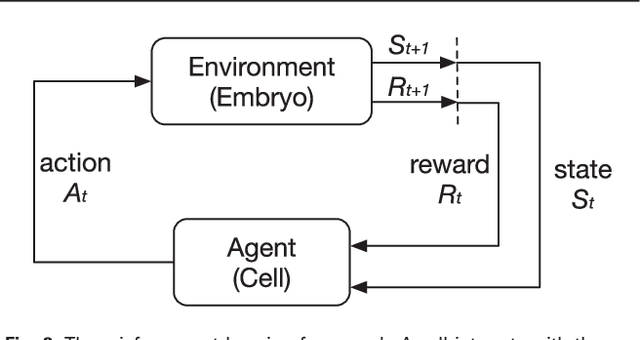
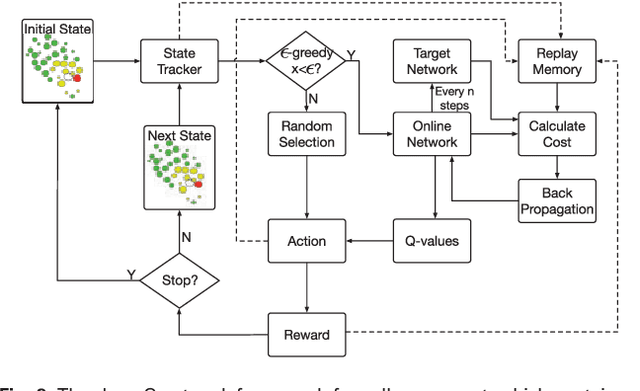
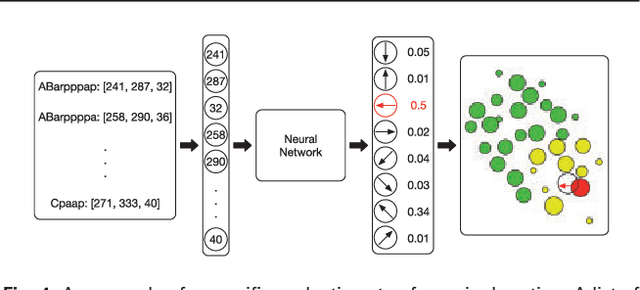
Abstract:Cell movement in the early phase of C. elegans development is regulated by a highly complex process in which a set of rules and connections are formulated at distinct scales. Previous efforts have shown that agent-based, multi-scale modeling systems can integrate physical and biological rules and provide new avenues to study developmental systems. However, the application of these systems to model cell movement is still challenging and requires a comprehensive understanding of regulation networks at the right scales. Recent developments in deep learning and reinforcement learning provide an unprecedented opportunity to explore cell movement using 3D time-lapse images. We present a deep reinforcement learning approach within an ABM system to characterize cell movement in C. elegans embryogenesis. Our modeling system captures the complexity of cell movement patterns in the embryo and overcomes the local optimization problem encountered by traditional rule-based, ABM that uses greedy algorithms. We tested our model with two real developmental processes: the anterior movement of the Cpaaa cell via intercalation and the rearrangement of the left-right asymmetry. In the first case, model results showed that Cpaaa's intercalation is an active directional cell movement caused by the continuous effects from a longer distance, as opposed to a passive movement caused by neighbor cell movements. This is because the learning-based simulation found that a passive movement model could not lead Cpaaa to the predefined destination. In the second case, a leader-follower mechanism well explained the collective cell movement pattern. These results showed that our approach to introduce deep reinforcement learning into ABM can test regulatory mechanisms by exploring cell migration paths in a reverse engineering perspective. This model opens new doors to explore large datasets generated by live imaging.
* We revised the manuscript to make it clearer to follow. Please notice that the Abstract shown in this page is slightly different than that in the manuscript due to the limitation of 1920 characters in arxiv.org
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge