Qinglei Cao
Toward Capturing Genetic Epistasis From Multivariate Genome-Wide Association Studies Using Mixed-Precision Kernel Ridge Regression
Sep 03, 2024



Abstract:We exploit the widening margin in tensor-core performance between [FP64/FP32/FP16/INT8,FP64/FP32/FP16/FP8/INT8] on NVIDIA [Ampere,Hopper] GPUs to boost the performance of output accuracy-preserving mixed-precision computation of Genome-Wide Association Studies (GWAS) of 305K patients from the UK BioBank, the largest-ever GWAS cohort studied for genetic epistasis using a multivariate approach. Tile-centric adaptive-precision linear algebraic techniques motivated by reducing data motion gain enhanced significance with low-precision GPU arithmetic. At the core of Kernel Ridge Regression (KRR) techniques for GWAS lie compute-bound cubic-complexity matrix operations that inhibit scaling to aspirational dimensions of the population, genotypes, and phenotypes. We accelerate KRR matrix generation by redesigning the computation for Euclidean distances to engage INT8 tensor cores while exploiting symmetry.We accelerate solution of the regularized KRR systems by deploying a new four-precision Cholesky-based solver, which, at 1.805 mixed-precision ExaOp/s on a nearly full Alps system, outperforms the state-of-the-art CPU-only REGENIE GWAS software by five orders of magnitude.
S3LLM: Large-Scale Scientific Software Understanding with LLMs using Source, Metadata, and Document
Mar 15, 2024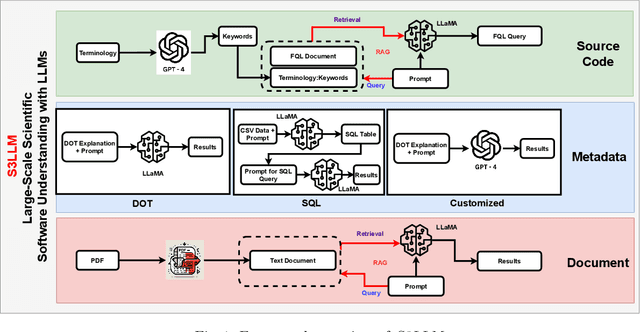
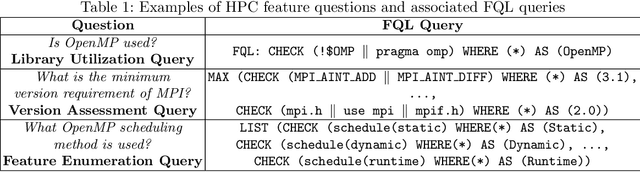
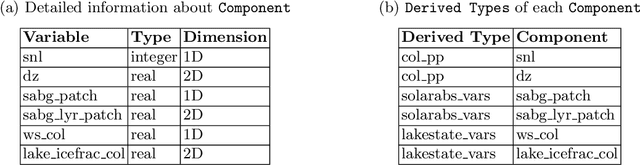
Abstract:The understanding of large-scale scientific software poses significant challenges due to its diverse codebase, extensive code length, and target computing architectures. The emergence of generative AI, specifically large language models (LLMs), provides novel pathways for understanding such complex scientific codes. This paper presents S3LLM, an LLM-based framework designed to enable the examination of source code, code metadata, and summarized information in conjunction with textual technical reports in an interactive, conversational manner through a user-friendly interface. S3LLM leverages open-source LLaMA-2 models to enhance code analysis through the automatic transformation of natural language queries into domain-specific language (DSL) queries. Specifically, it translates these queries into Feature Query Language (FQL), enabling efficient scanning and parsing of entire code repositories. In addition, S3LLM is equipped to handle diverse metadata types, including DOT, SQL, and customized formats. Furthermore, S3LLM incorporates retrieval augmented generation (RAG) and LangChain technologies to directly query extensive documents. S3LLM demonstrates the potential of using locally deployed open-source LLMs for the rapid understanding of large-scale scientific computing software, eliminating the need for extensive coding expertise, and thereby making the process more efficient and effective. S3LLM is available at https://github.com/ResponsibleAILab/s3llm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge