Zhirong Bao
Augmenting C. elegans Microscopic Dataset for Accelerated Pattern Recognition
May 31, 2019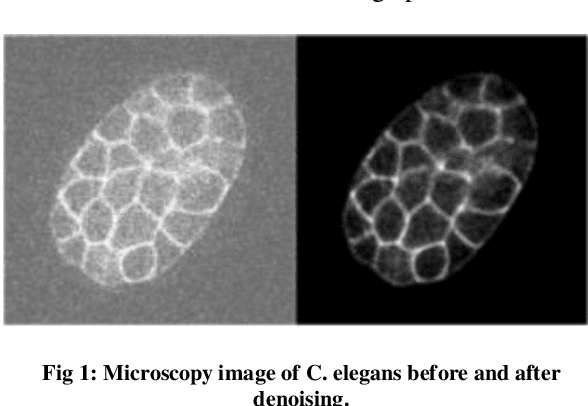
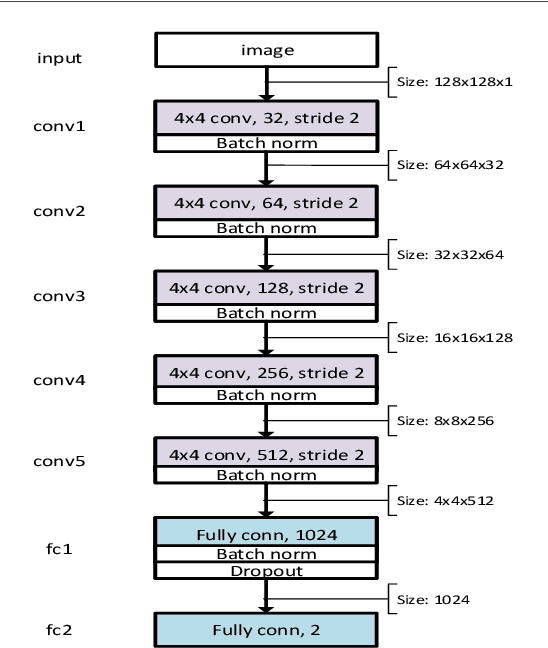
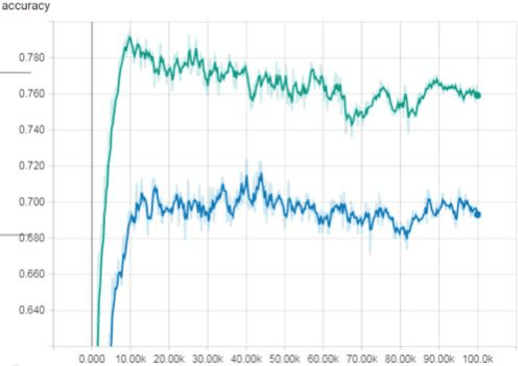
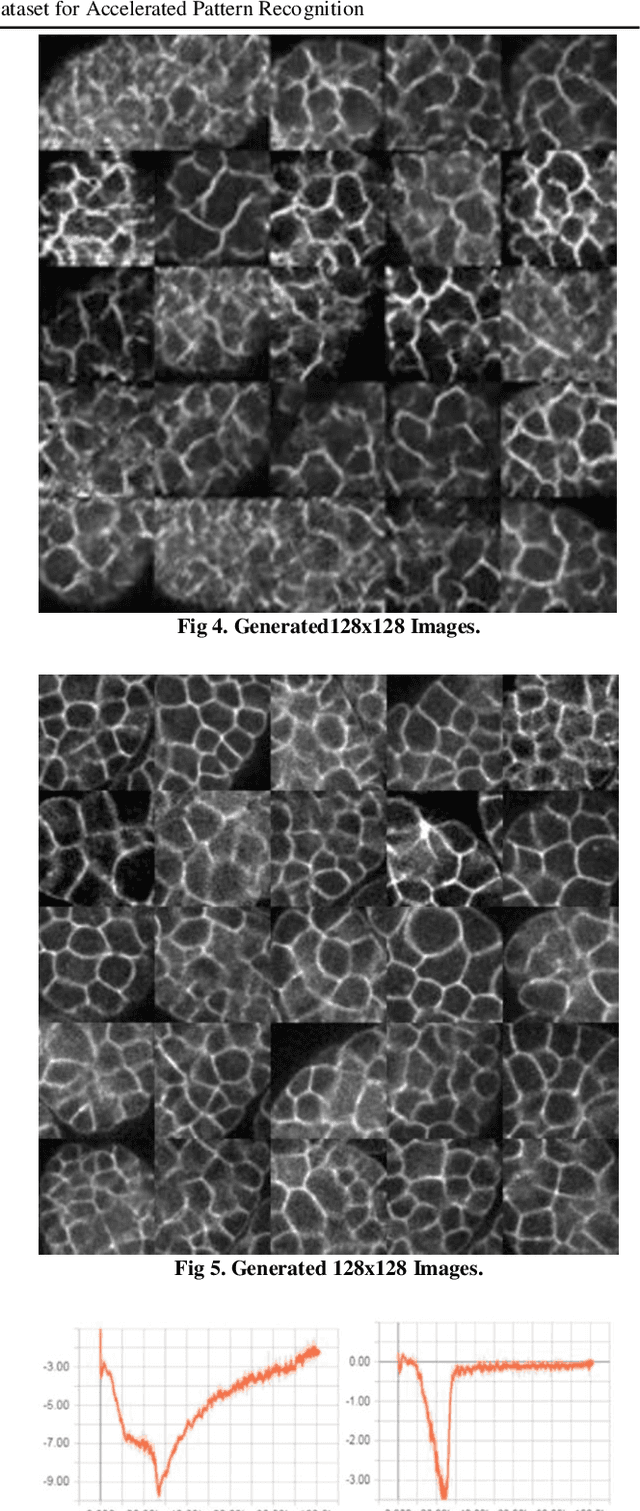
Abstract:The detection of cell shape changes in 3D time-lapse images of complex tissues is an important task. However, it is a challenging and tedious task to establish a comprehensive dataset to improve the performance of deep learning models. In the paper, we present a deep learning approach to augment 3D live images of the Caenorhabditis elegans embryo, so that we can further speed up the specific structural pattern recognition. We use an unsupervised training over unlabeled images to generate supplementary datasets for further pattern recognition. Technically, we used Alex-style neural networks in a generative adversarial network framework to generate new datasets that have common features of the C. elegans membrane structure. We also made the dataset available for a broad scientific community.
Deep Reinforcement Learning of Cell Movement in the Early Stage of C. elegans Embryogenesis
Mar 02, 2018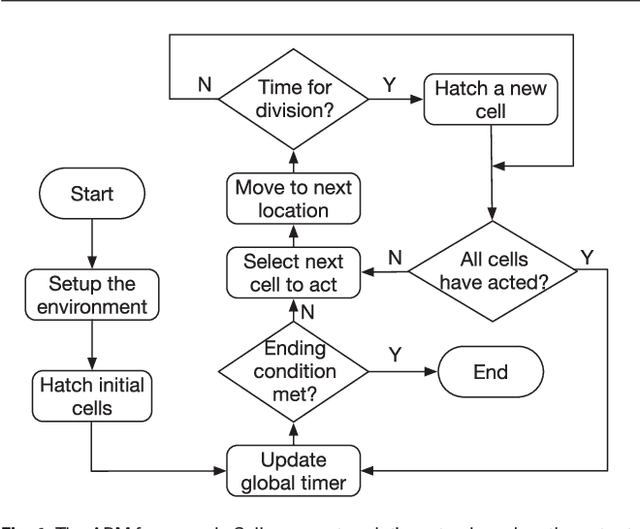
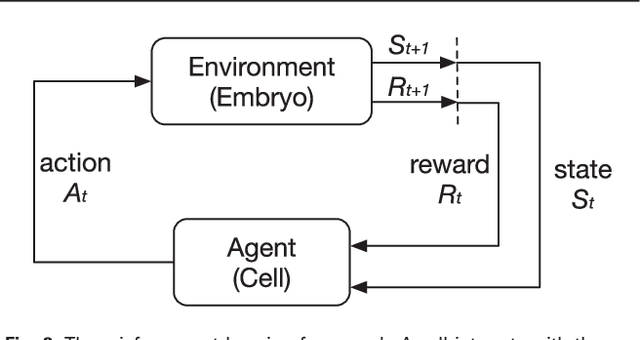
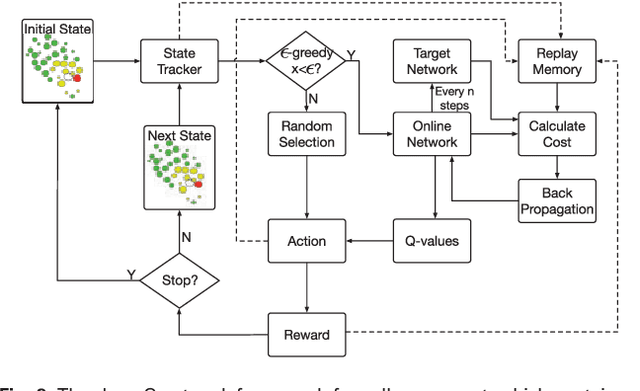
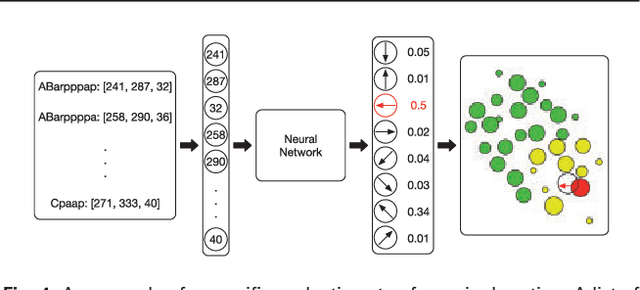
Abstract:Cell movement in the early phase of C. elegans development is regulated by a highly complex process in which a set of rules and connections are formulated at distinct scales. Previous efforts have shown that agent-based, multi-scale modeling systems can integrate physical and biological rules and provide new avenues to study developmental systems. However, the application of these systems to model cell movement is still challenging and requires a comprehensive understanding of regulation networks at the right scales. Recent developments in deep learning and reinforcement learning provide an unprecedented opportunity to explore cell movement using 3D time-lapse images. We present a deep reinforcement learning approach within an ABM system to characterize cell movement in C. elegans embryogenesis. Our modeling system captures the complexity of cell movement patterns in the embryo and overcomes the local optimization problem encountered by traditional rule-based, ABM that uses greedy algorithms. We tested our model with two real developmental processes: the anterior movement of the Cpaaa cell via intercalation and the rearrangement of the left-right asymmetry. In the first case, model results showed that Cpaaa's intercalation is an active directional cell movement caused by the continuous effects from a longer distance, as opposed to a passive movement caused by neighbor cell movements. This is because the learning-based simulation found that a passive movement model could not lead Cpaaa to the predefined destination. In the second case, a leader-follower mechanism well explained the collective cell movement pattern. These results showed that our approach to introduce deep reinforcement learning into ABM can test regulatory mechanisms by exploring cell migration paths in a reverse engineering perspective. This model opens new doors to explore large datasets generated by live imaging.
* We revised the manuscript to make it clearer to follow. Please notice that the Abstract shown in this page is slightly different than that in the manuscript due to the limitation of 1920 characters in arxiv.org
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge