Calvin Chen
The Weather Paradox: Why Precipitation Fails to Predict Traffic Accident Severity in Large-Scale US Data
Jan 01, 2026Abstract:This study investigates the predictive capacity of environmental, temporal, and spatial factors on traffic accident severity in the United States. Using a dataset of 500,000 U.S. traffic accidents spanning 2016-2023, we trained an XGBoost classifier optimized through randomized search cross-validation and adjusted for class imbalance via class weighting. The final model achieves an overall accuracy of 78%, with strong performance on the majority class (Severity 2), attaining 87% precision and recall. Feature importance analysis reveals that time of day, geographic location, and weather-related variables, including visibility, temperature, and wind speed, rank among the strongest predictors of accident severity. However, contrary to initial hypotheses, precipitation and visibility demonstrate limited predictive power, potentially reflecting behavioral adaptation by drivers under overtly hazardous conditions. The dataset's predominance of mid-level severity accidents constrains the model's capacity to learn meaningful patterns for extreme cases, highlighting the need for alternative sampling strategies, enhanced feature engineering, and integration of external datasets. These findings contribute to evidence-based traffic management and suggest future directions for severity prediction research.
TrackingMiM: Efficient Mamba-in-Mamba Serialization for Real-time UAV Object Tracking
Jul 02, 2025Abstract:The Vision Transformer (ViT) model has long struggled with the challenge of quadratic complexity, a limitation that becomes especially critical in unmanned aerial vehicle (UAV) tracking systems, where data must be processed in real time. In this study, we explore the recently proposed State-Space Model, Mamba, leveraging its computational efficiency and capability for long-sequence modeling to effectively process dense image sequences in tracking tasks. First, we highlight the issue of temporal inconsistency in existing Mamba-based methods, specifically the failure to account for temporal continuity in the Mamba scanning mechanism. Secondly, building upon this insight,we propose TrackingMiM, a Mamba-in-Mamba architecture, a minimal-computation burden model for handling image sequence of tracking problem. In our framework, the mamba scan is performed in a nested way while independently process temporal and spatial coherent patch tokens. While the template frame is encoded as query token and utilized for tracking in every scan. Extensive experiments conducted on five UAV tracking benchmarks confirm that the proposed TrackingMiM achieves state-of-the-art precision while offering noticeable higher speed in UAV tracking.
MambaControl: Anatomy Graph-Enhanced Mamba ControlNet with Fourier Refinement for Diffusion-Based Disease Trajectory Prediction
May 15, 2025Abstract:Modelling disease progression in precision medicine requires capturing complex spatio-temporal dynamics while preserving anatomical integrity. Existing methods often struggle with longitudinal dependencies and structural consistency in progressive disorders. To address these limitations, we introduce MambaControl, a novel framework that integrates selective state-space modelling with diffusion processes for high-fidelity prediction of medical image trajectories. To better capture subtle structural changes over time while maintaining anatomical consistency, MambaControl combines Mamba-based long-range modelling with graph-guided anatomical control to more effectively represent anatomical correlations. Furthermore, we introduce Fourier-enhanced spectral graph representations to capture spatial coherence and multiscale detail, enabling MambaControl to achieve state-of-the-art performance in Alzheimer's disease prediction. Quantitative and regional evaluations demonstrate improved progression prediction quality and anatomical fidelity, highlighting its potential for personalised prognosis and clinical decision support.
TextInPlace: Indoor Visual Place Recognition in Repetitive Structures with Scene Text Spotting and Verification
Mar 09, 2025Abstract:Visual Place Recognition (VPR) is a crucial capability for long-term autonomous robots, enabling them to identify previously visited locations using visual information. However, existing methods remain limited in indoor settings due to the highly repetitive structures inherent in such environments. We observe that scene text typically appears in indoor spaces, serving to distinguish visually similar but different places. This inspires us to propose TextInPlace, a simple yet effective VPR framework that integrates Scene Text Spotting (STS) to mitigate visual perceptual ambiguity in repetitive indoor environments. Specifically, TextInPlace adopts a dual-branch architecture within a local parameter sharing network. The VPR branch employs attention-based aggregation to extract global descriptors for coarse-grained retrieval, while the STS branch utilizes a bridging text spotter to detect and recognize scene text. Finally, the discriminative text is filtered to compute text similarity and re-rank the top-K retrieved images. To bridge the gap between current text-based repetitive indoor scene datasets and the typical scenarios encountered in robot navigation, we establish an indoor VPR benchmark dataset, called Maze-with-Text. Extensive experiments on both custom and public datasets demonstrate that TextInPlace achieves superior performance over existing methods that rely solely on appearance information. The dataset, code, and trained models are publicly available at https://github.com/HqiTao/TextInPlace.
Generating near-infrared facial expression datasets with dimensional affect labels
Jun 28, 2022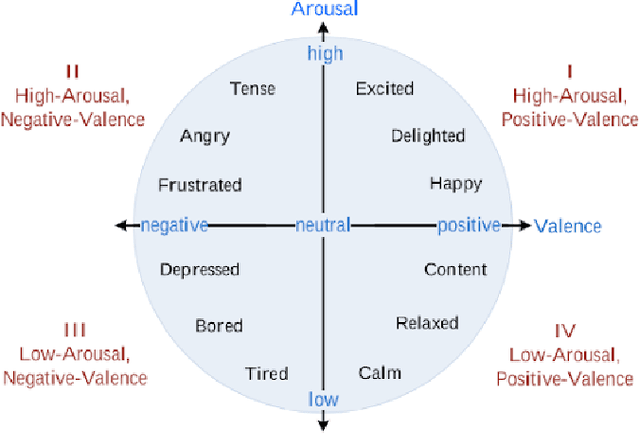
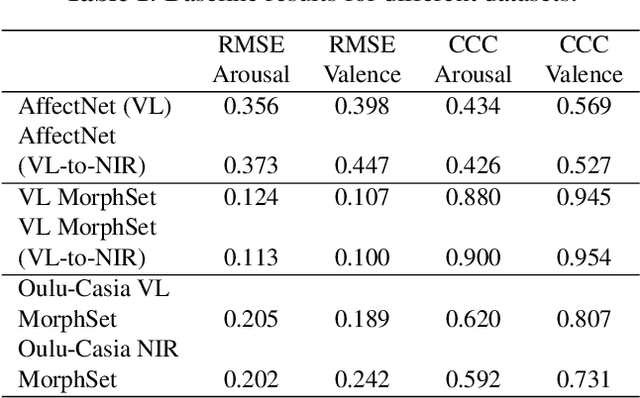
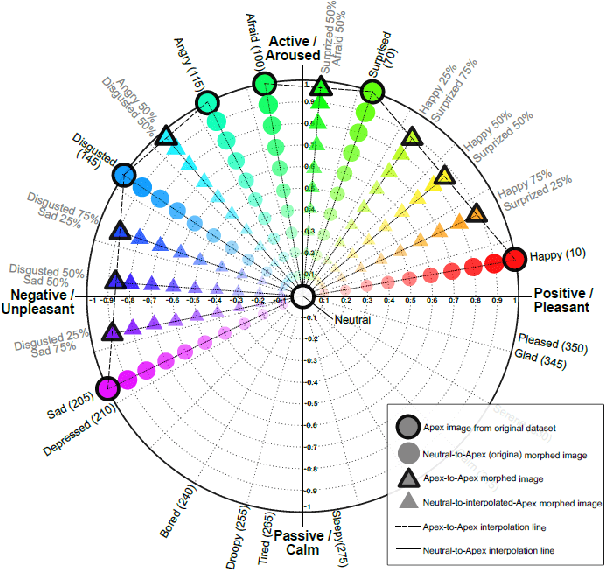

Abstract:Facial expression analysis has long been an active research area of computer vision. Traditional methods mainly analyse images for prototypical discrete emotions; as a result, they do not provide an accurate depiction of the complex emotional states in humans. Furthermore, illumination variance remains a challenge for face analysis in the visible light spectrum. To address these issues, we propose using a dimensional model based on valence and arousal to represent a wider range of emotions, in combination with near infra-red (NIR) imagery, which is more robust to illumination changes. Since there are no existing NIR facial expression datasets with valence-arousal labels available, we present two complementary data augmentation methods (face morphing and CycleGAN approach) to create NIR image datasets with dimensional emotion labels from existing categorical and/or visible-light datasets. Our experiments show that these generated NIR datasets are comparable to existing datasets in terms of data quality and baseline prediction performance.
Clinical Named Entity Recognition using Contextualized Token Representations
Jun 23, 2021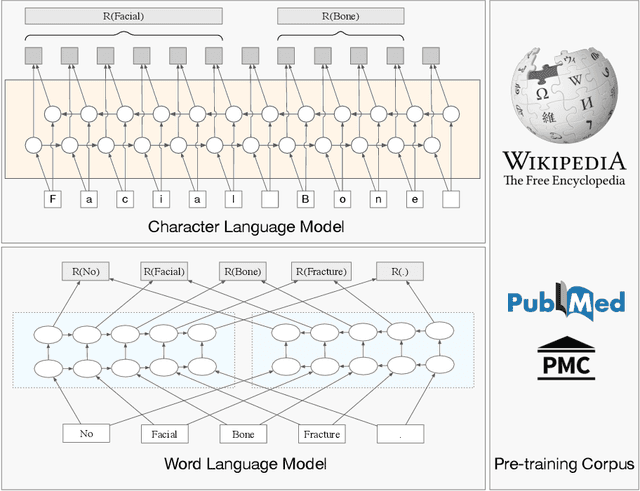
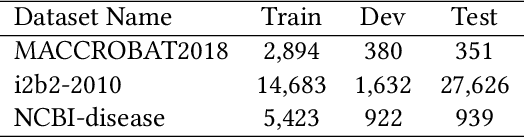
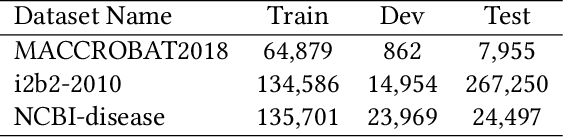

Abstract:The clinical named entity recognition (CNER) task seeks to locate and classify clinical terminologies into predefined categories, such as diagnostic procedure, disease disorder, severity, medication, medication dosage, and sign symptom. CNER facilitates the study of side-effect on medications including identification of novel phenomena and human-focused information extraction. Existing approaches in extracting the entities of interests focus on using static word embeddings to represent each word. However, one word can have different interpretations that depend on the context of the sentences. Evidently, static word embeddings are insufficient to integrate the diverse interpretation of a word. To overcome this challenge, the technique of contextualized word embedding has been introduced to better capture the semantic meaning of each word based on its context. Two of these language models, ELMo and Flair, have been widely used in the field of Natural Language Processing to generate the contextualized word embeddings on domain-generic documents. However, these embeddings are usually too general to capture the proximity among vocabularies of specific domains. To facilitate various downstream applications using clinical case reports (CCRs), we pre-train two deep contextualized language models, Clinical Embeddings from Language Model (C-ELMo) and Clinical Contextual String Embeddings (C-Flair) using the clinical-related corpus from the PubMed Central. Explicit experiments show that our models gain dramatic improvements compared to both static word embeddings and domain-generic language models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge