Peipei Ping
Platform for Representation and Integration of multimodal Molecular Embeddings
Jul 10, 2025Abstract:Existing machine learning methods for molecular (e.g., gene) embeddings are restricted to specific tasks or data modalities, limiting their effectiveness within narrow domains. As a result, they fail to capture the full breadth of gene functions and interactions across diverse biological contexts. In this study, we have systematically evaluated knowledge representations of biomolecules across multiple dimensions representing a task-agnostic manner spanning three major data sources, including omics experimental data, literature-derived text data, and knowledge graph-based representations. To distinguish between meaningful biological signals from chance correlations, we devised an adjusted variant of Singular Vector Canonical Correlation Analysis (SVCCA) that quantifies signal redundancy and complementarity across different data modalities and sources. These analyses reveal that existing embeddings capture largely non-overlapping molecular signals, highlighting the value of embedding integration. Building on this insight, we propose Platform for Representation and Integration of multimodal Molecular Embeddings (PRISME), a machine learning based workflow using an autoencoder to integrate these heterogeneous embeddings into a unified multimodal representation. We validated this approach across various benchmark tasks, where PRISME demonstrated consistent performance, and outperformed individual embedding methods in missing value imputations. This new framework supports comprehensive modeling of biomolecules, advancing the development of robust, broadly applicable multimodal embeddings optimized for downstream biomedical machine learning applications.
Atherosclerosis through Hierarchical Explainable Neural Network Analysis
Jul 10, 2025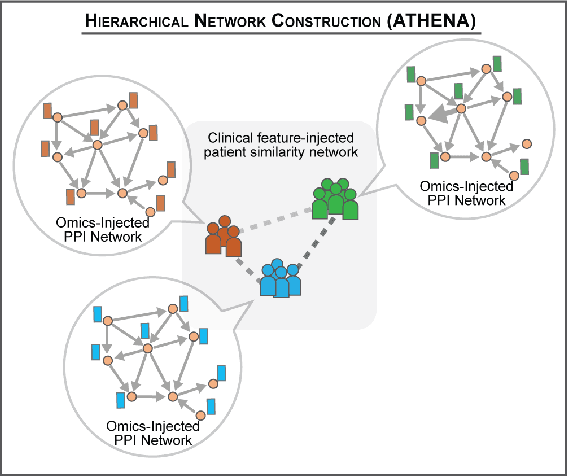
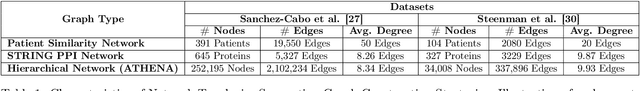
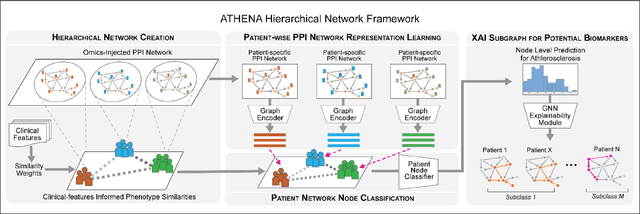
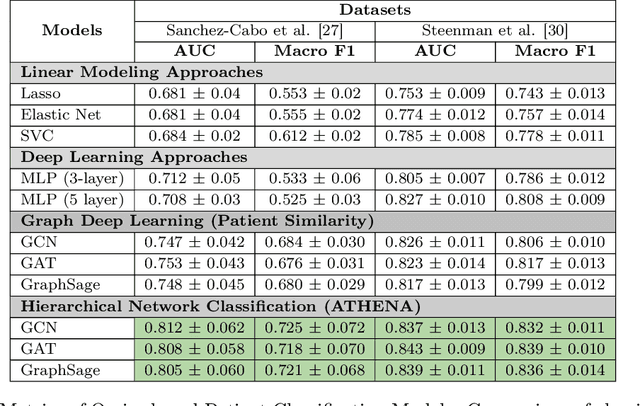
Abstract:In this work, we study the problem pertaining to personalized classification of subclinical atherosclerosis by developing a hierarchical graph neural network framework to leverage two characteristic modalities of a patient: clinical features within the context of the cohort, and molecular data unique to individual patients. Current graph-based methods for disease classification detect patient-specific molecular fingerprints, but lack consistency and comprehension regarding cohort-wide features, which are an essential requirement for understanding pathogenic phenotypes across diverse atherosclerotic trajectories. Furthermore, understanding patient subtypes often considers clinical feature similarity in isolation, without integration of shared pathogenic interdependencies among patients. To address these challenges, we introduce ATHENA: Atherosclerosis Through Hierarchical Explainable Neural Network Analysis, which constructs a novel hierarchical network representation through integrated modality learning; subsequently, it optimizes learned patient-specific molecular fingerprints that reflect individual omics data, enforcing consistency with cohort-wide patterns. With a primary clinical dataset of 391 patients, we demonstrate that this heterogeneous alignment of clinical features with molecular interaction patterns has significantly boosted subclinical atherosclerosis classification performance across various baselines by up to 13% in area under the receiver operating curve (AUC) and 20% in F1 score. Taken together, ATHENA enables mechanistically-informed patient subtype discovery through explainable AI (XAI)-driven subnetwork clustering; this novel integration framework strengthens personalized intervention strategies, thereby improving the prediction of atherosclerotic disease progression and management of their clinical actionable outcomes.
Bridge2AI: Building A Cross-disciplinary Curriculum Towards AI-Enhanced Biomedical and Clinical Care
May 20, 2025Abstract:Objective: As AI becomes increasingly central to healthcare, there is a pressing need for bioinformatics and biomedical training systems that are personalized and adaptable. Materials and Methods: The NIH Bridge2AI Training, Recruitment, and Mentoring (TRM) Working Group developed a cross-disciplinary curriculum grounded in collaborative innovation, ethical data stewardship, and professional development within an adapted Learning Health System (LHS) framework. Results: The curriculum integrates foundational AI modules, real-world projects, and a structured mentee-mentor network spanning Bridge2AI Grand Challenges and the Bridge Center. Guided by six learner personas, the program tailors educational pathways to individual needs while supporting scalability. Discussion: Iterative refinement driven by continuous feedback ensures that content remains responsive to learner progress and emerging trends. Conclusion: With over 30 scholars and 100 mentors engaged across North America, the TRM model demonstrates how adaptive, persona-informed training can build interdisciplinary competencies and foster an integrative, ethically grounded AI education in biomedical contexts.
Building an Ethical and Trustworthy Biomedical AI Ecosystem for the Translational and Clinical Integration of Foundational Models
Jul 18, 2024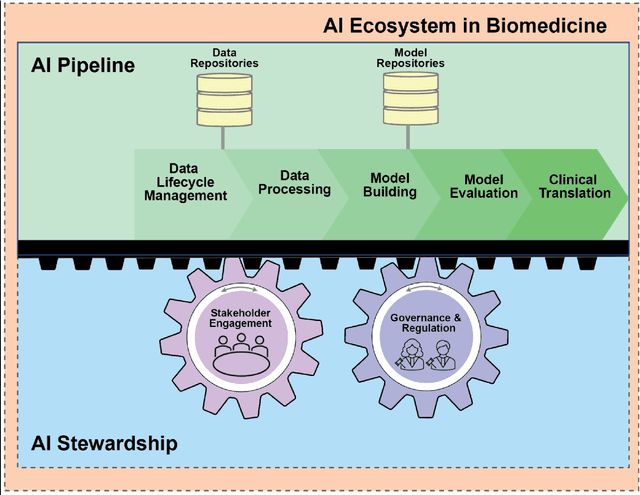
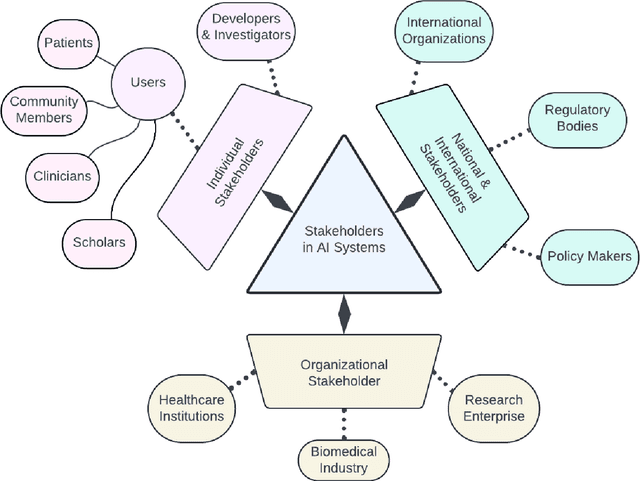
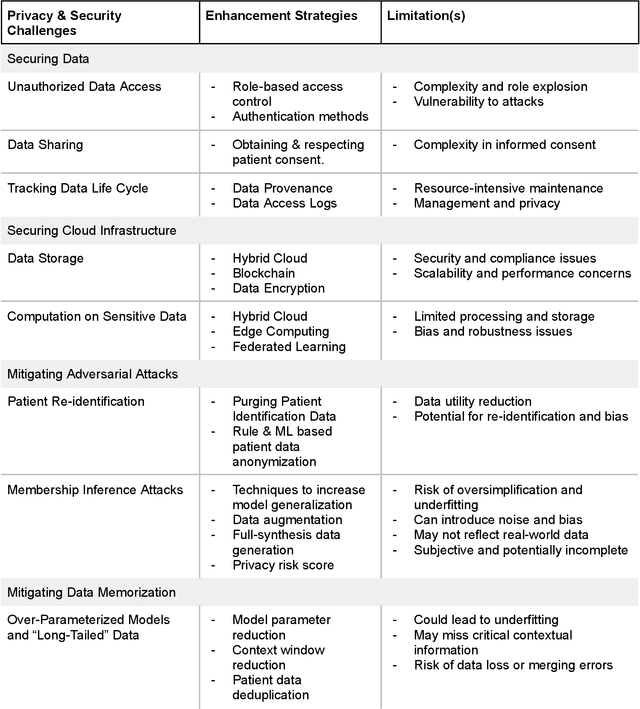
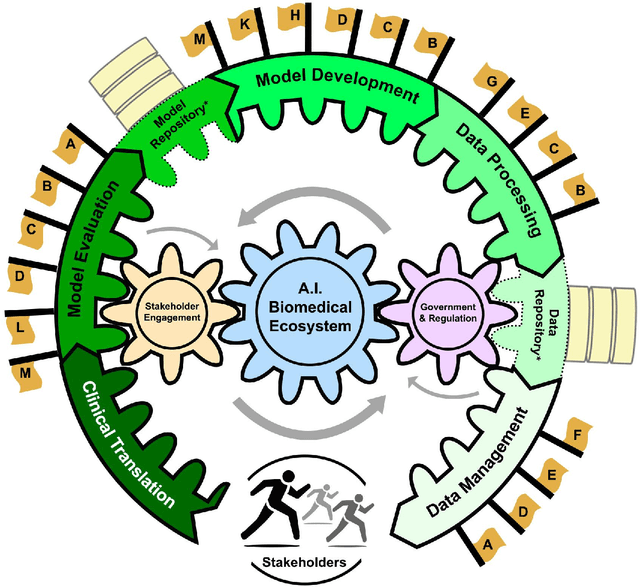
Abstract:Foundational Models (FMs) are emerging as the cornerstone of the biomedical AI ecosystem due to their ability to represent and contextualize multimodal biomedical data. These capabilities allow FMs to be adapted for various tasks, including biomedical reasoning, hypothesis generation, and clinical decision-making. This review paper examines the foundational components of an ethical and trustworthy AI (ETAI) biomedical ecosystem centered on FMs, highlighting key challenges and solutions. The ETAI biomedical ecosystem is defined by seven key components which collectively integrate FMs into clinical settings: Data Lifecycle Management, Data Processing, Model Development, Model Evaluation, Clinical Translation, AI Governance and Regulation, and Stakeholder Engagement. While the potential of biomedical AI is immense, it requires heightened ethical vigilance and responsibility. For instance, biases can arise from data, algorithms, and user interactions, necessitating techniques to assess and mitigate bias prior to, during, and after model development. Moreover, interpretability, explainability, and accountability are key to ensuring the trustworthiness of AI systems, while workflow transparency in training, testing, and evaluation is crucial for reproducibility. Safeguarding patient privacy and security involves addressing challenges in data access, cloud data privacy, patient re-identification, membership inference attacks, and data memorization. Additionally, AI governance and regulation are essential for ethical AI use in biomedicine, guided by global standards. Furthermore, stakeholder engagement is essential at every stage of the AI pipeline and lifecycle for clinical translation. By adhering to these principles, we can harness the transformative potential of AI and develop an ETAI ecosystem.
Explainable Biomedical Hypothesis Generation via Retrieval Augmented Generation enabled Large Language Models
Jul 17, 2024Abstract:The vast amount of biomedical information available today presents a significant challenge for investigators seeking to digest, process, and understand these findings effectively. Large Language Models (LLMs) have emerged as powerful tools to navigate this complex and challenging data landscape. However, LLMs may lead to hallucinatory responses, making Retrieval Augmented Generation (RAG) crucial for achieving accurate information. In this protocol, we present RUGGED (Retrieval Under Graph-Guided Explainable disease Distinction), a comprehensive workflow designed to support investigators with knowledge integration and hypothesis generation, identifying validated paths forward. Relevant biomedical information from publications and knowledge bases are reviewed, integrated, and extracted via text-mining association analysis and explainable graph prediction models on disease nodes, forecasting potential links among drugs and diseases. These analyses, along with biomedical texts, are integrated into a framework that facilitates user-directed mechanism elucidation as well as hypothesis exploration through RAG-enabled LLMs. A clinical use-case demonstrates RUGGED's ability to evaluate and recommend therapeutics for Arrhythmogenic Cardiomyopathy (ACM) and Dilated Cardiomyopathy (DCM), analyzing prescribed drugs for molecular interactions and unexplored uses. The platform minimizes LLM hallucinations, offers actionable insights, and improves the investigation of novel therapeutics.
CliBench: Multifaceted Evaluation of Large Language Models in Clinical Decisions on Diagnoses, Procedures, Lab Tests Orders and Prescriptions
Jun 14, 2024



Abstract:The integration of Artificial Intelligence (AI), especially Large Language Models (LLMs), into the clinical diagnosis process offers significant potential to improve the efficiency and accessibility of medical care. While LLMs have shown some promise in the medical domain, their application in clinical diagnosis remains underexplored, especially in real-world clinical practice, where highly sophisticated, patient-specific decisions need to be made. Current evaluations of LLMs in this field are often narrow in scope, focusing on specific diseases or specialties and employing simplified diagnostic tasks. To bridge this gap, we introduce CliBench, a novel benchmark developed from the MIMIC IV dataset, offering a comprehensive and realistic assessment of LLMs' capabilities in clinical diagnosis. This benchmark not only covers diagnoses from a diverse range of medical cases across various specialties but also incorporates tasks of clinical significance: treatment procedure identification, lab test ordering and medication prescriptions. Supported by structured output ontologies, CliBench enables a precise and multi-granular evaluation, offering an in-depth understanding of LLM's capability on diverse clinical tasks of desired granularity. We conduct a zero-shot evaluation of leading LLMs to assess their proficiency in clinical decision-making. Our preliminary results shed light on the potential and limitations of current LLMs in clinical settings, providing valuable insights for future advancements in LLM-powered healthcare.
Know2BIO: A Comprehensive Dual-View Benchmark for Evolving Biomedical Knowledge Graphs
Oct 05, 2023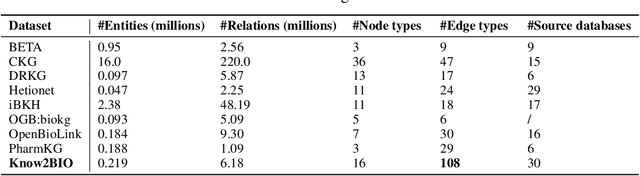
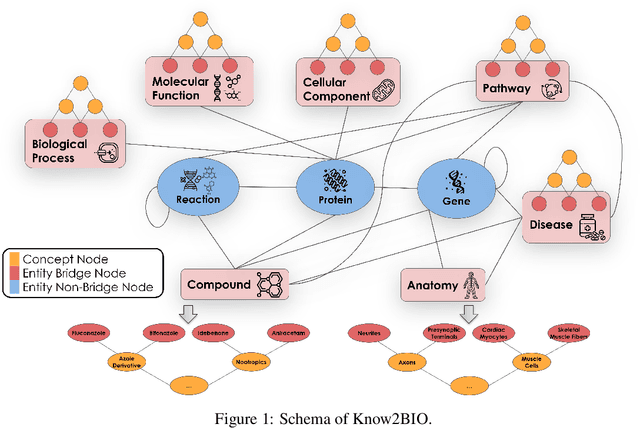
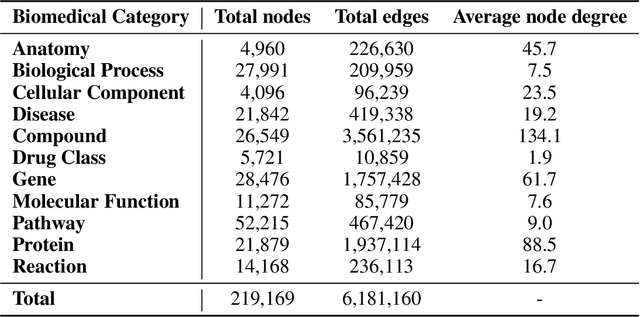
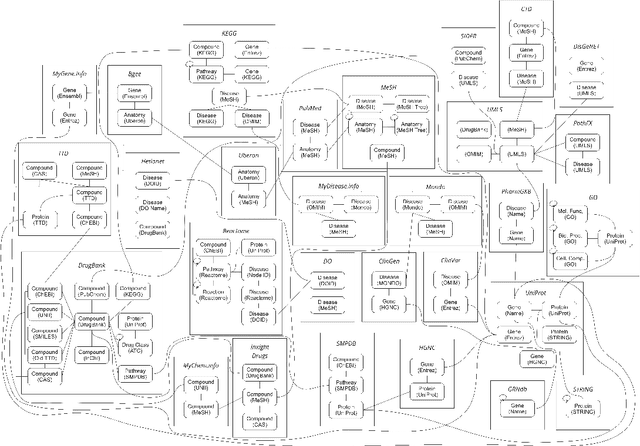
Abstract:Knowledge graphs (KGs) have emerged as a powerful framework for representing and integrating complex biomedical information. However, assembling KGs from diverse sources remains a significant challenge in several aspects, including entity alignment, scalability, and the need for continuous updates to keep pace with scientific advancements. Moreover, the representative power of KGs is often limited by the scarcity of multi-modal data integration. To overcome these challenges, we propose Know2BIO, a general-purpose heterogeneous KG benchmark for the biomedical domain. Know2BIO integrates data from 30 diverse sources, capturing intricate relationships across 11 biomedical categories. It currently consists of ~219,000 nodes and ~6,200,000 edges. Know2BIO is capable of user-directed automated updating to reflect the latest knowledge in biomedical science. Furthermore, Know2BIO is accompanied by multi-modal data: node features including text descriptions, protein and compound sequences and structures, enabling the utilization of emerging natural language processing methods and multi-modal data integration strategies. We evaluate KG representation models on Know2BIO, demonstrating its effectiveness as a benchmark for KG representation learning in the biomedical field. Data and source code of Know2BIO are available at https://github.com/Yijia-Xiao/Know2BIO/.
Clinical Named Entity Recognition using Contextualized Token Representations
Jun 23, 2021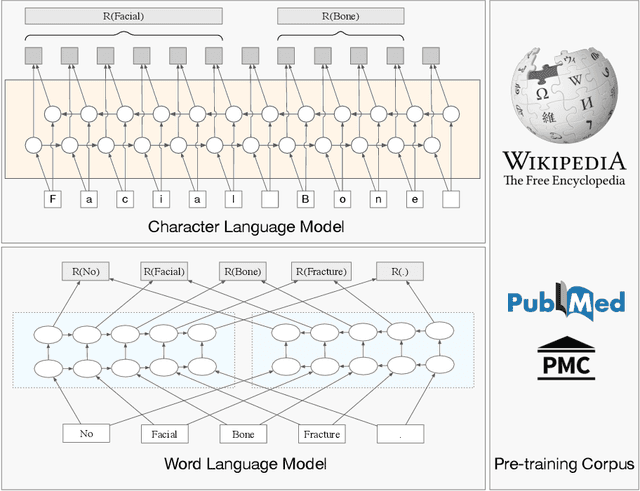
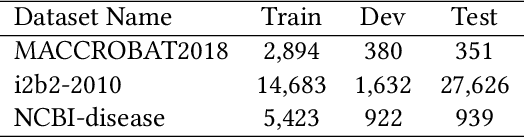
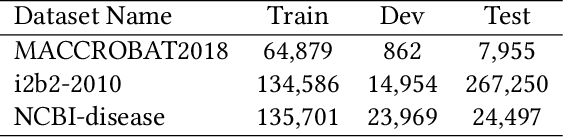

Abstract:The clinical named entity recognition (CNER) task seeks to locate and classify clinical terminologies into predefined categories, such as diagnostic procedure, disease disorder, severity, medication, medication dosage, and sign symptom. CNER facilitates the study of side-effect on medications including identification of novel phenomena and human-focused information extraction. Existing approaches in extracting the entities of interests focus on using static word embeddings to represent each word. However, one word can have different interpretations that depend on the context of the sentences. Evidently, static word embeddings are insufficient to integrate the diverse interpretation of a word. To overcome this challenge, the technique of contextualized word embedding has been introduced to better capture the semantic meaning of each word based on its context. Two of these language models, ELMo and Flair, have been widely used in the field of Natural Language Processing to generate the contextualized word embeddings on domain-generic documents. However, these embeddings are usually too general to capture the proximity among vocabularies of specific domains. To facilitate various downstream applications using clinical case reports (CCRs), we pre-train two deep contextualized language models, Clinical Embeddings from Language Model (C-ELMo) and Clinical Contextual String Embeddings (C-Flair) using the clinical-related corpus from the PubMed Central. Explicit experiments show that our models gain dramatic improvements compared to both static word embeddings and domain-generic language models.
CREATe: Clinical Report Extraction and Annotation Technology
Feb 28, 2021



Abstract:Clinical case reports are written descriptions of the unique aspects of a particular clinical case, playing an essential role in sharing clinical experiences about atypical disease phenotypes and new therapies. However, to our knowledge, there has been no attempt to develop an end-to-end system to annotate, index, or otherwise curate these reports. In this paper, we propose a novel computational resource platform, CREATe, for extracting, indexing, and querying the contents of clinical case reports. CREATe fosters an environment of sustainable resource support and discovery, enabling researchers to overcome the challenges of information science. An online video of the demonstration can be viewed at https://youtu.be/Q8owBQYTjDc.
Clinical Temporal Relation Extraction with Probabilistic Soft Logic Regularization and Global Inference
Dec 16, 2020



Abstract:There has been a steady need in the medical community to precisely extract the temporal relations between clinical events. In particular, temporal information can facilitate a variety of downstream applications such as case report retrieval and medical question answering. Existing methods either require expensive feature engineering or are incapable of modeling the global relational dependencies among the events. In this paper, we propose a novel method, Clinical Temporal ReLation Exaction with Probabilistic Soft Logic Regularization and Global Inference (CTRL-PG) to tackle the problem at the document level. Extensive experiments on two benchmark datasets, I2B2-2012 and TB-Dense, demonstrate that CTRL-PG significantly outperforms baseline methods for temporal relation extraction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge