Karol Watson
Atherosclerosis through Hierarchical Explainable Neural Network Analysis
Jul 10, 2025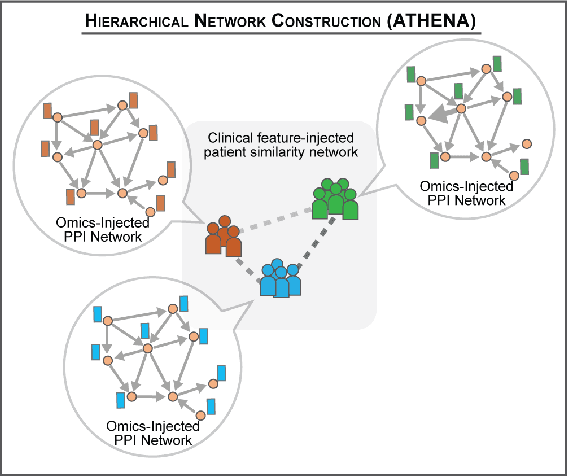
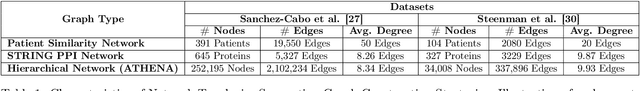
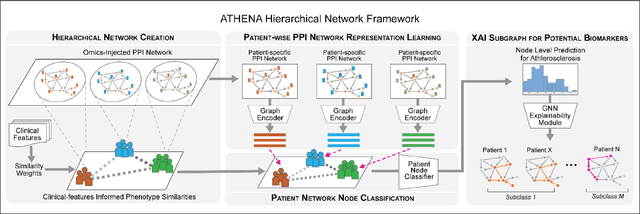
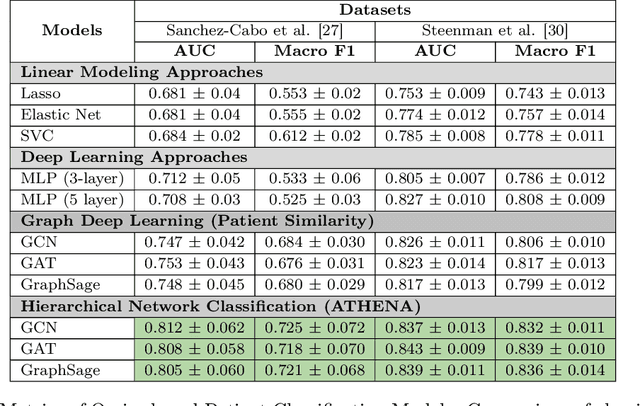
Abstract:In this work, we study the problem pertaining to personalized classification of subclinical atherosclerosis by developing a hierarchical graph neural network framework to leverage two characteristic modalities of a patient: clinical features within the context of the cohort, and molecular data unique to individual patients. Current graph-based methods for disease classification detect patient-specific molecular fingerprints, but lack consistency and comprehension regarding cohort-wide features, which are an essential requirement for understanding pathogenic phenotypes across diverse atherosclerotic trajectories. Furthermore, understanding patient subtypes often considers clinical feature similarity in isolation, without integration of shared pathogenic interdependencies among patients. To address these challenges, we introduce ATHENA: Atherosclerosis Through Hierarchical Explainable Neural Network Analysis, which constructs a novel hierarchical network representation through integrated modality learning; subsequently, it optimizes learned patient-specific molecular fingerprints that reflect individual omics data, enforcing consistency with cohort-wide patterns. With a primary clinical dataset of 391 patients, we demonstrate that this heterogeneous alignment of clinical features with molecular interaction patterns has significantly boosted subclinical atherosclerosis classification performance across various baselines by up to 13% in area under the receiver operating curve (AUC) and 20% in F1 score. Taken together, ATHENA enables mechanistically-informed patient subtype discovery through explainable AI (XAI)-driven subnetwork clustering; this novel integration framework strengthens personalized intervention strategies, thereby improving the prediction of atherosclerotic disease progression and management of their clinical actionable outcomes.
Building an Ethical and Trustworthy Biomedical AI Ecosystem for the Translational and Clinical Integration of Foundational Models
Jul 18, 2024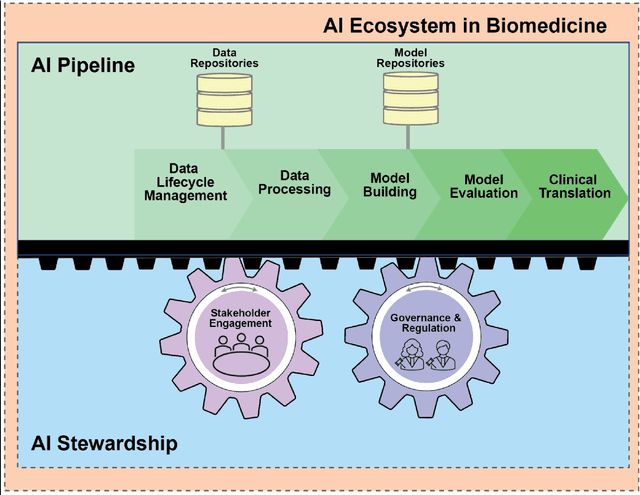
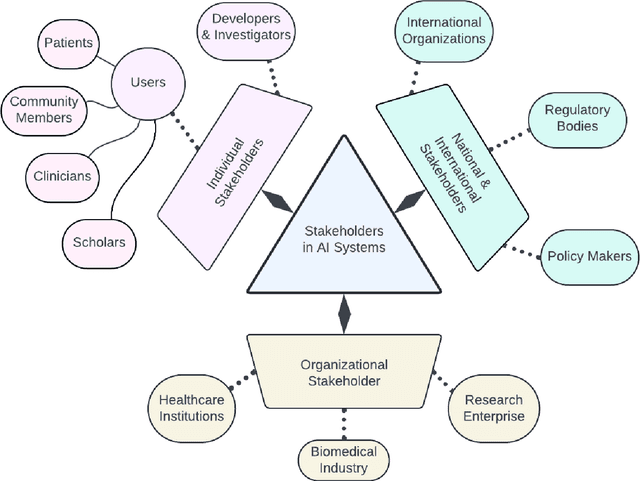
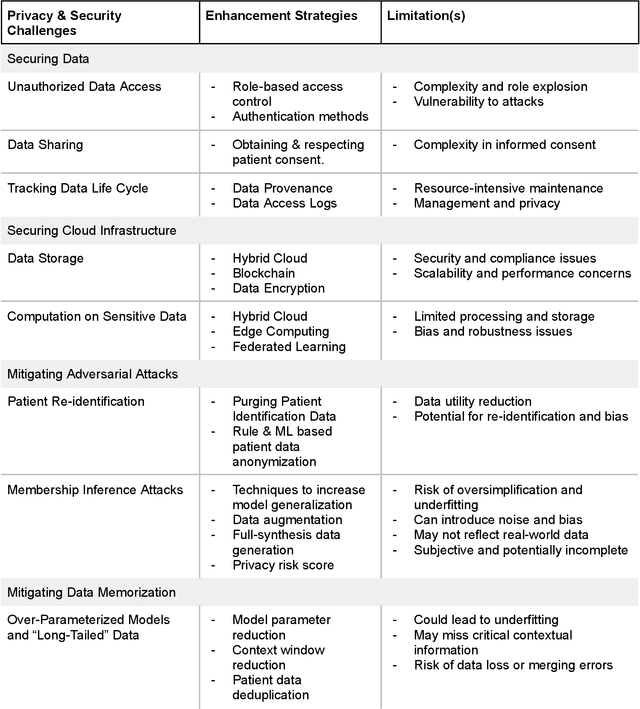
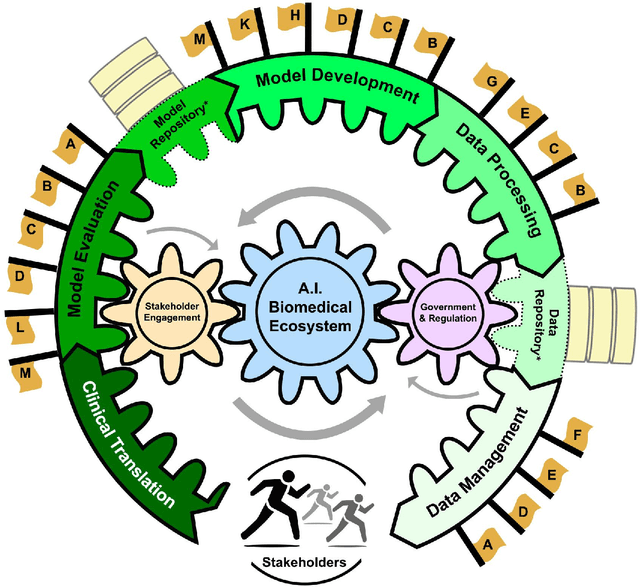
Abstract:Foundational Models (FMs) are emerging as the cornerstone of the biomedical AI ecosystem due to their ability to represent and contextualize multimodal biomedical data. These capabilities allow FMs to be adapted for various tasks, including biomedical reasoning, hypothesis generation, and clinical decision-making. This review paper examines the foundational components of an ethical and trustworthy AI (ETAI) biomedical ecosystem centered on FMs, highlighting key challenges and solutions. The ETAI biomedical ecosystem is defined by seven key components which collectively integrate FMs into clinical settings: Data Lifecycle Management, Data Processing, Model Development, Model Evaluation, Clinical Translation, AI Governance and Regulation, and Stakeholder Engagement. While the potential of biomedical AI is immense, it requires heightened ethical vigilance and responsibility. For instance, biases can arise from data, algorithms, and user interactions, necessitating techniques to assess and mitigate bias prior to, during, and after model development. Moreover, interpretability, explainability, and accountability are key to ensuring the trustworthiness of AI systems, while workflow transparency in training, testing, and evaluation is crucial for reproducibility. Safeguarding patient privacy and security involves addressing challenges in data access, cloud data privacy, patient re-identification, membership inference attacks, and data memorization. Additionally, AI governance and regulation are essential for ethical AI use in biomedicine, guided by global standards. Furthermore, stakeholder engagement is essential at every stage of the AI pipeline and lifecycle for clinical translation. By adhering to these principles, we can harness the transformative potential of AI and develop an ETAI ecosystem.
Unsupervised Airway Tree Clustering with Deep Learning: The Multi-Ethnic Study of Atherosclerosis (MESA) Lung Study
Feb 28, 2024


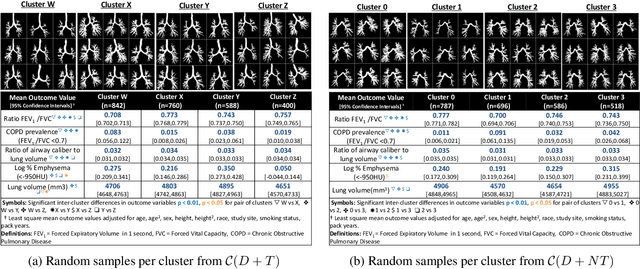
Abstract:High-resolution full lung CT scans now enable the detailed segmentation of airway trees up to the 6th branching generation. The airway binary masks display very complex tree structures that may encode biological information relevant to disease risk and yet remain challenging to exploit via traditional methods such as meshing or skeletonization. Recent clinical studies suggest that some variations in shape patterns and caliber of the human airway tree are highly associated with adverse health outcomes, including all-cause mortality and incident COPD. However, quantitative characterization of variations observed on CT segmented airway tree remain incomplete, as does our understanding of the clinical and developmental implications of such. In this work, we present an unsupervised deep-learning pipeline for feature extraction and clustering of human airway trees, learned directly from projections of 3D airway segmentations. We identify four reproducible and clinically distinct airway sub-types in the MESA Lung CT cohort.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge