Bogdan J. Matuszewski
CVAM-Pose: Conditional Variational Autoencoder for Multi-Object Monocular Pose Estimation
Oct 11, 2024



Abstract:Estimating rigid objects' poses is one of the fundamental problems in computer vision, with a range of applications across automation and augmented reality. Most existing approaches adopt one network per object class strategy, depend heavily on objects' 3D models, depth data, and employ a time-consuming iterative refinement, which could be impractical for some applications. This paper presents a novel approach, CVAM-Pose, for multi-object monocular pose estimation that addresses these limitations. The CVAM-Pose method employs a label-embedded conditional variational autoencoder network, to implicitly abstract regularised representations of multiple objects in a single low-dimensional latent space. This autoencoding process uses only images captured by a projective camera and is robust to objects' occlusion and scene clutter. The classes of objects are one-hot encoded and embedded throughout the network. The proposed label-embedded pose regression strategy interprets the learnt latent space representations utilising continuous pose representations. Ablation tests and systematic evaluations demonstrate the scalability and efficiency of the CVAM-Pose method for multi-object scenarios. The proposed CVAM-Pose outperforms competing latent space approaches. For example, it is respectively 25% and 20% better than AAE and Multi-Path methods, when evaluated using the $\mathrm{AR_{VSD}}$ metric on the Linemod-Occluded dataset. It also achieves results somewhat comparable to methods reliant on 3D models reported in BOP challenges. Code available: https://github.com/JZhao12/CVAM-Pose
Polyp Segmentation Generalisability of Pretrained Backbones
May 24, 2024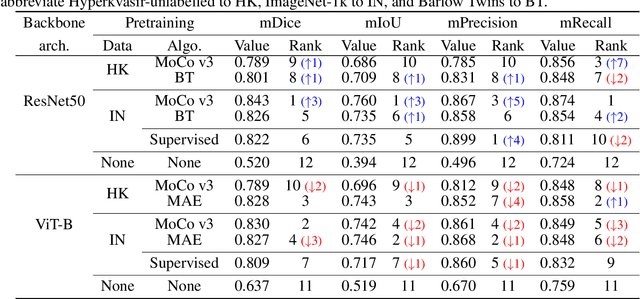
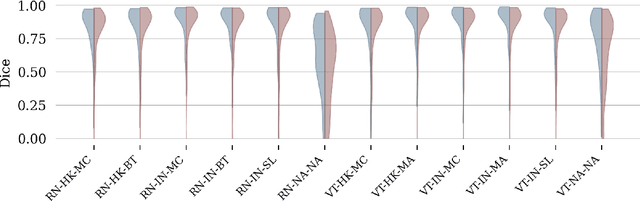
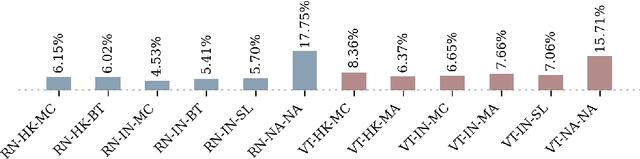
Abstract:It has recently been demonstrated that pretraining backbones in a self-supervised manner generally provides better fine-tuned polyp segmentation performance, and that models with ViT-B backbones typically perform better than models with ResNet50 backbones. In this paper, we extend this recent work to consider generalisability. I.e., we assess the performance of models on a different dataset to that used for fine-tuning, accounting for variation in network architecture and pretraining pipeline (algorithm and dataset). This reveals how well models with different pretrained backbones generalise to data of a somewhat different distribution to the training data, which will likely arise in deployment due to different cameras and demographics of patients, amongst other factors. We observe that the previous findings, regarding pretraining pipelines for polyp segmentation, hold true when considering generalisability. However, our results imply that models with ResNet50 backbones typically generalise better, despite being outperformed by models with ViT-B backbones in evaluation on the test set from the same dataset used for fine-tuning.
A Study on Self-Supervised Pretraining for Vision Problems in Gastrointestinal Endoscopy
Jan 11, 2024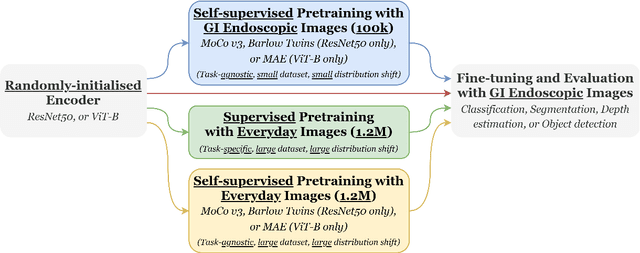
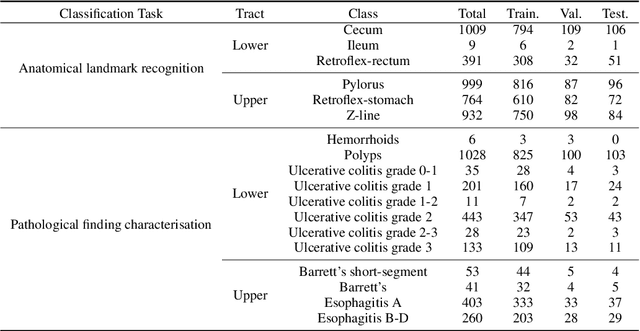
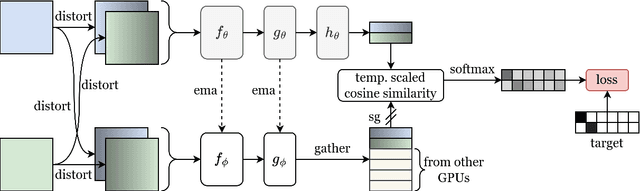
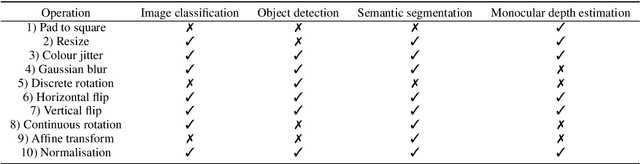
Abstract:Solutions to vision tasks in gastrointestinal endoscopy (GIE) conventionally use image encoders pretrained in a supervised manner with ImageNet-1k as backbones. However, the use of modern self-supervised pretraining algorithms and a recent dataset of 100k unlabelled GIE images (Hyperkvasir-unlabelled) may allow for improvements. In this work, we study the fine-tuned performance of models with ResNet50 and ViT-B backbones pretrained in self-supervised and supervised manners with ImageNet-1k and Hyperkvasir-unlabelled (self-supervised only) in a range of GIE vision tasks. In addition to identifying the most suitable pretraining pipeline and backbone architecture for each task, out of those considered, our results suggest: that self-supervised pretraining generally produces more suitable backbones for GIE vision tasks than supervised pretraining; that self-supervised pretraining with ImageNet-1k is typically more suitable than pretraining with Hyperkvasir-unlabelled, with the notable exception of monocular depth estimation in colonoscopy; and that ViT-Bs are more suitable in polyp segmentation and monocular depth estimation in colonoscopy, ResNet50s are more suitable in polyp detection, and both architectures perform similarly in anatomical landmark recognition and pathological finding characterisation. We hope this work draws attention to the complexity of pretraining for GIE vision tasks, informs this development of more suitable approaches than the convention, and inspires further research on this topic to help advance this development. Code available: \underline{github.com/ESandML/SSL4GIE}
SimCol3D -- 3D Reconstruction during Colonoscopy Challenge
Jul 20, 2023Abstract:Colorectal cancer is one of the most common cancers in the world. While colonoscopy is an effective screening technique, navigating an endoscope through the colon to detect polyps is challenging. A 3D map of the observed surfaces could enhance the identification of unscreened colon tissue and serve as a training platform. However, reconstructing the colon from video footage remains unsolved due to numerous factors such as self-occlusion, reflective surfaces, lack of texture, and tissue deformation that limit feature-based methods. Learning-based approaches hold promise as robust alternatives, but necessitate extensive datasets. By establishing a benchmark, the 2022 EndoVis sub-challenge SimCol3D aimed to facilitate data-driven depth and pose prediction during colonoscopy. The challenge was hosted as part of MICCAI 2022 in Singapore. Six teams from around the world and representatives from academia and industry participated in the three sub-challenges: synthetic depth prediction, synthetic pose prediction, and real pose prediction. This paper describes the challenge, the submitted methods, and their results. We show that depth prediction in virtual colonoscopy is robustly solvable, while pose estimation remains an open research question.
FCN-Transformer Feature Fusion for Polyp Segmentation
Aug 17, 2022Abstract:Colonoscopy is widely recognised as the gold standard procedure for the early detection of colorectal cancer (CRC). Segmentation is valuable for two significant clinical applications, namely lesion detection and classification, providing means to improve accuracy and robustness. The manual segmentation of polyps in colonoscopy images is time-consuming. As a result, the use of deep learning (DL) for automation of polyp segmentation has become important. However, DL-based solutions can be vulnerable to overfitting and the resulting inability to generalise to images captured by different colonoscopes. Recent transformer-based architectures for semantic segmentation both achieve higher performance and generalise better than alternatives, however typically predict a segmentation map of $\frac{h}{4}\times\frac{w}{4}$ spatial dimensions for a $h\times w$ input image. To this end, we propose a new architecture for full-size segmentation which leverages the strengths of a transformer in extracting the most important features for segmentation in a primary branch, while compensating for its limitations in full-size prediction with a secondary fully convolutional branch. The resulting features from both branches are then fused for final prediction of a $h\times w$ segmentation map. We demonstrate our method's state-of-the-art performance with respect to the mDice, mIoU, mPrecision, and mRecall metrics, on both the Kvasir-SEG and CVC-ClinicDB dataset benchmarks. Additionally, we train the model on each of these datasets and evaluate on the other to demonstrate its superior generalisation performance.
* 16 pages, 4 figures
mREAL-GAN: Generating Multiple Residential Electrical Appliance Load Profiles with Inter-Dependencies using a Generative Adversarial Network
Dec 08, 2021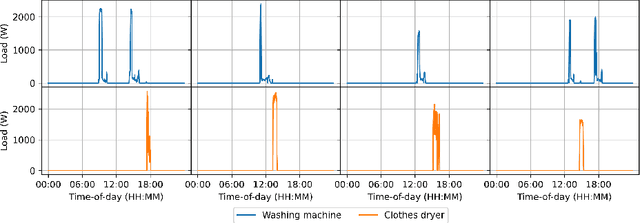
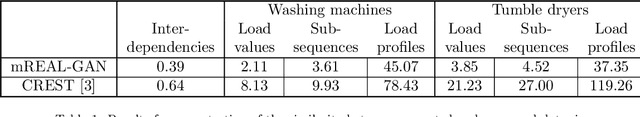
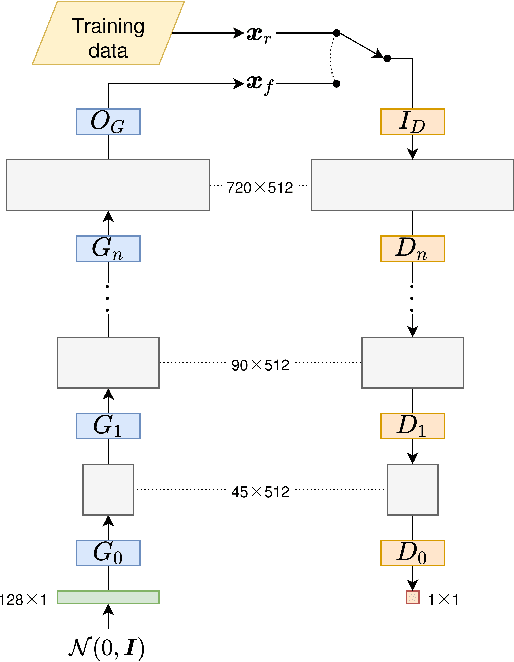
Abstract:In this paper, we introduce mREAL-GAN, a generative adversarial network (GAN) for the parallel generation of multiple residential electrical appliance load (mREAL) profiles. mREAL-GAN is intended for use in community-scale low-voltage network analysis, and represents a departure from previous methods for this purpose, which break the generation of appliance load profiles into several steps and largely model each appliance independently. Instead, mREAL-GAN models appliance load profiles in an end-to-end manner, and generates multiple appliance load profiles in parallel in a way that captures inter-dependencies. We show that mREAL-GAN generates load profiles for individual appliance-types with greater fidelity than a popular example of previous methods, and demonstrate its ability to capture inter-dependencies between appliances.
Gland Segmentation in Colon Histology Images: The GlaS Challenge Contest
Sep 01, 2016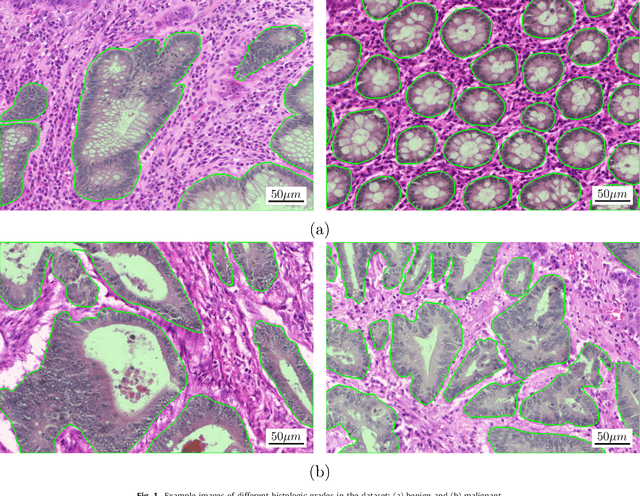
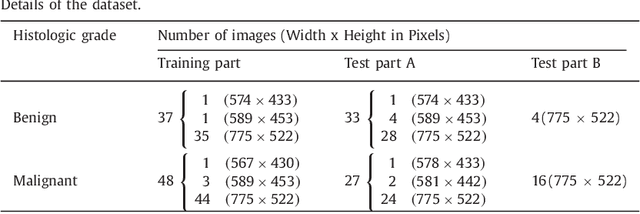
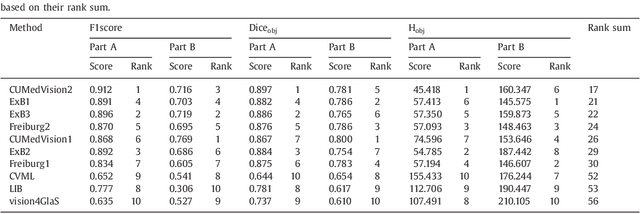
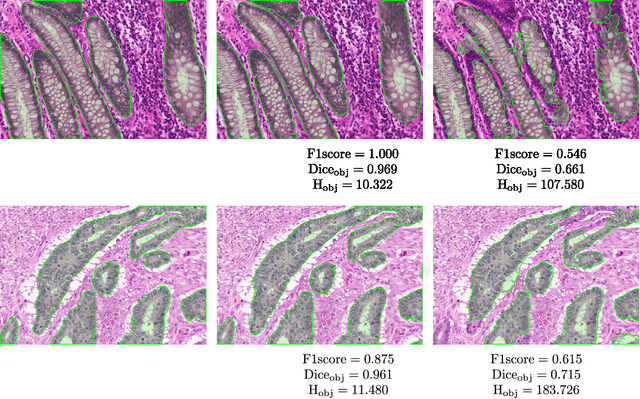
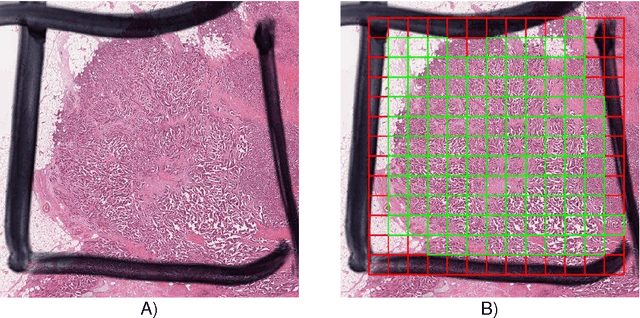
Abstract:Colorectal adenocarcinoma originating in intestinal glandular structures is the most common form of colon cancer. In clinical practice, the morphology of intestinal glands, including architectural appearance and glandular formation, is used by pathologists to inform prognosis and plan the treatment of individual patients. However, achieving good inter-observer as well as intra-observer reproducibility of cancer grading is still a major challenge in modern pathology. An automated approach which quantifies the morphology of glands is a solution to the problem. This paper provides an overview to the Gland Segmentation in Colon Histology Images Challenge Contest (GlaS) held at MICCAI'2015. Details of the challenge, including organization, dataset and evaluation criteria, are presented, along with the method descriptions and evaluation results from the top performing methods.
Assessment of algorithms for mitosis detection in breast cancer histopathology images
Nov 21, 2014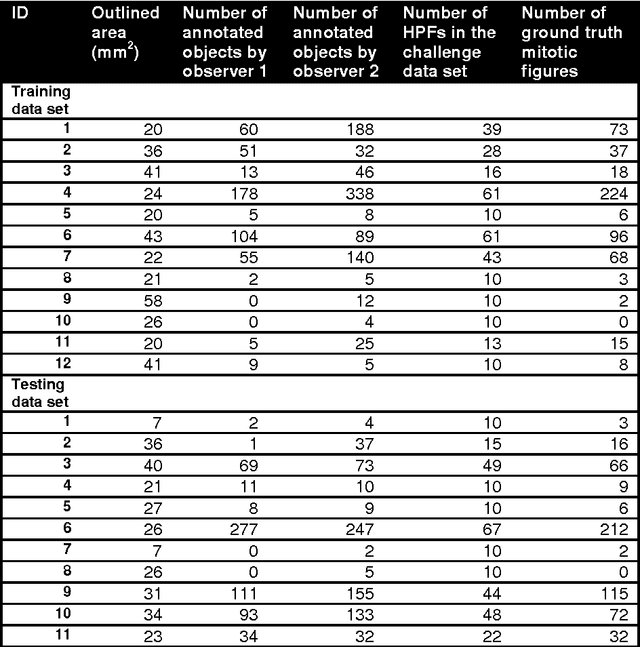
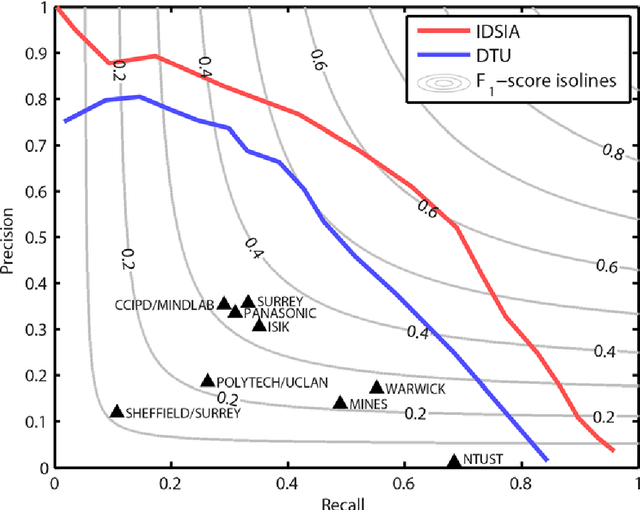
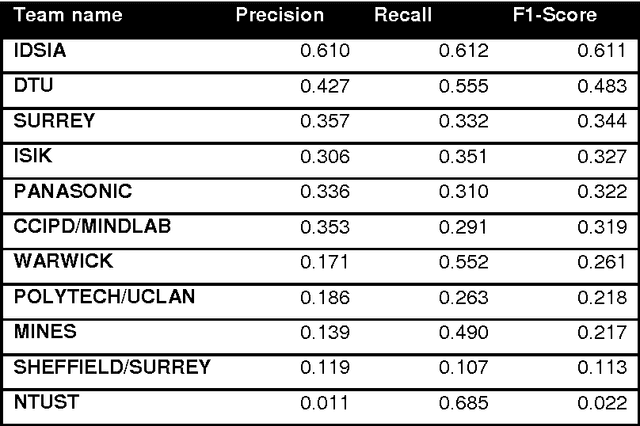
Abstract:The proliferative activity of breast tumors, which is routinely estimated by counting of mitotic figures in hematoxylin and eosin stained histology sections, is considered to be one of the most important prognostic markers. However, mitosis counting is laborious, subjective and may suffer from low inter-observer agreement. With the wider acceptance of whole slide images in pathology labs, automatic image analysis has been proposed as a potential solution for these issues. In this paper, the results from the Assessment of Mitosis Detection Algorithms 2013 (AMIDA13) challenge are described. The challenge was based on a data set consisting of 12 training and 11 testing subjects, with more than one thousand annotated mitotic figures by multiple observers. Short descriptions and results from the evaluation of eleven methods are presented. The top performing method has an error rate that is comparable to the inter-observer agreement among pathologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge