Javier Morlana
Topological SLAM in colonoscopies leveraging deep features and topological priors
Sep 25, 2024Abstract:We introduce ColonSLAM, a system that combines classical multiple-map metric SLAM with deep features and topological priors to create topological maps of the whole colon. The SLAM pipeline by itself is able to create disconnected individual metric submaps representing locations from short video subsections of the colon, but is not able to merge covisible submaps due to deformations and the limited performance of the SIFT descriptor in the medical domain. ColonSLAM is guided by topological priors and combines a deep localization network trained to distinguish if two images come from the same place or not and the soft verification of a transformer-based matching network, being able to relate far-in-time submaps during an exploration, grouping them in nodes imaging the same colon place, building more complex maps than any other approach in the literature. We demonstrate our approach in the Endomapper dataset, showing its potential for producing maps of the whole colon in real human explorations. Code and models are available at: https://github.com/endomapper/ColonSLAM.
SimCol3D -- 3D Reconstruction during Colonoscopy Challenge
Jul 20, 2023Abstract:Colorectal cancer is one of the most common cancers in the world. While colonoscopy is an effective screening technique, navigating an endoscope through the colon to detect polyps is challenging. A 3D map of the observed surfaces could enhance the identification of unscreened colon tissue and serve as a training platform. However, reconstructing the colon from video footage remains unsolved due to numerous factors such as self-occlusion, reflective surfaces, lack of texture, and tissue deformation that limit feature-based methods. Learning-based approaches hold promise as robust alternatives, but necessitate extensive datasets. By establishing a benchmark, the 2022 EndoVis sub-challenge SimCol3D aimed to facilitate data-driven depth and pose prediction during colonoscopy. The challenge was hosted as part of MICCAI 2022 in Singapore. Six teams from around the world and representatives from academia and industry participated in the three sub-challenges: synthetic depth prediction, synthetic pose prediction, and real pose prediction. This paper describes the challenge, the submitted methods, and their results. We show that depth prediction in virtual colonoscopy is robustly solvable, while pose estimation remains an open research question.
ColonMapper: topological mapping and localization for colonoscopy
May 09, 2023Abstract:Mapping and localization in endoluminal cavities from colonoscopies or gastroscopies has to overcome the challenge of significant shape and illumination changes between reobservations of the same endoluminal location. Instead of geometrical maps that strongly rely on a fixed scene geometry, topological maps are more adequate because they focus on visual place recognition, i.e. the capability to determine if two video shots are imaging the same location. We propose a topological mapping and localization system able to operate on real human colonoscopies. The map is a graph where each node codes a colon location by a set of real images of that location. The edges represent traversability between two nodes. For close-in-time images, where scene changes are minor, place recognition can be successfully managed with the recent transformers-based image-matching algorithms. However, under long-term changes -- such as different colonoscopies of the same patient -- feature-based matching fails. To address this, we propose a GeM global descriptor able to achieve high recall with significant changes in the scene. The addition of a Bayesian filter processing the map graph boosts the accuracy of the long-term place recognition, enabling relocalization in a previously built map. In the experiments, we construct a map during the withdrawal phase of a first colonoscopy. Subsequently, we prove the ability to relocalize within this map during a second colonoscopy of the same patient two weeks later. Code and models will be available upon acceptance.
EndoMapper dataset of complete calibrated endoscopy procedures
Apr 29, 2022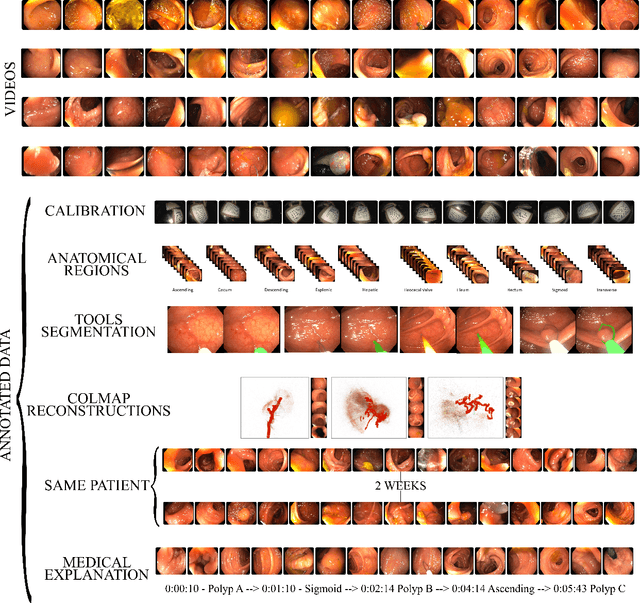
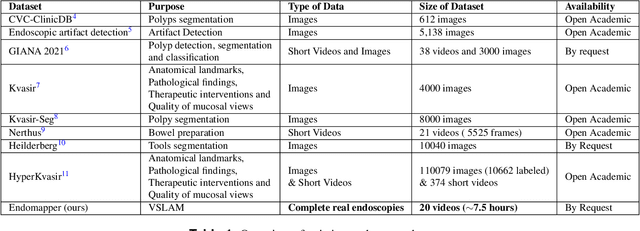

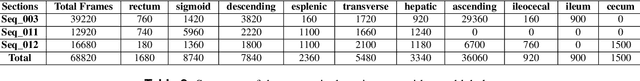
Abstract:Computer-assisted systems are becoming broadly used in medicine. In endoscopy, most research focuses on automatic detection of polyps or other pathologies, but localization and navigation of the endoscope is completely performed manually by physicians. To broaden this research and bring spatial Artificial Intelligence to endoscopies, data from complete procedures are needed. This data will be used to build a 3D mapping and localization systems that can perform special task like, for example, detect blind zones during exploration, provide automatic polyp measurements, guide doctors to a polyp found in a previous exploration and retrieve previous images of the same area aligning them for easy comparison. These systems will provide an improvement in the quality and precision of the procedures while lowering the burden on the physicians. This paper introduces the Endomapper dataset, the first collection of complete endoscopy sequences acquired during regular medical practice, including slow and careful screening explorations, making secondary use of medical data. Its original purpose is to facilitate the development and evaluation of VSLAM (Visual Simultaneous Localization and Mapping) methods in real endoscopy data. The first release of the dataset is composed of 59 sequences with more than 15 hours of video. It is also the first endoscopic dataset that includes both the computed geometric and photometric endoscope calibration with the original calibration videos. Meta-data and annotations associated to the dataset varies from anatomical landmark and description of the procedure labeling, tools segmentation masks, COLMAP 3D reconstructions, simulated sequences with groundtruth and meta-data related to special cases, such as sequences from the same patient. This information will improve the research in endoscopic VSLAM, as well as other research lines, and create new research lines.
Reuse your features: unifying retrieval and feature-metric alignment
Apr 13, 2022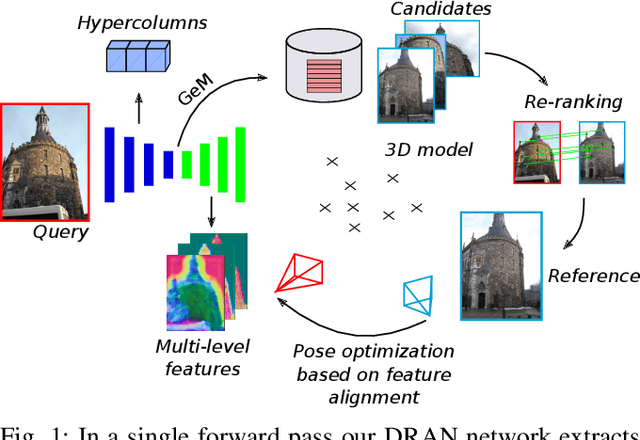
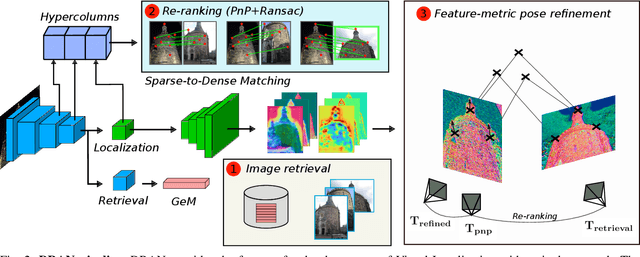
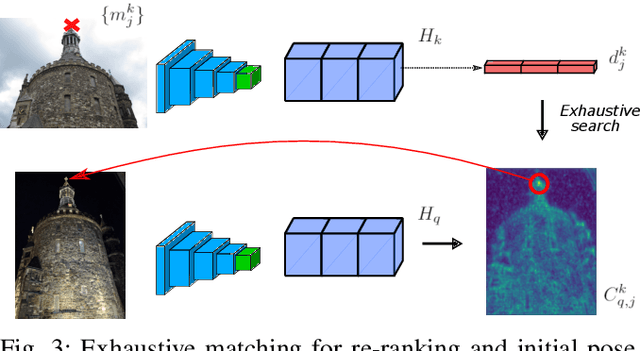
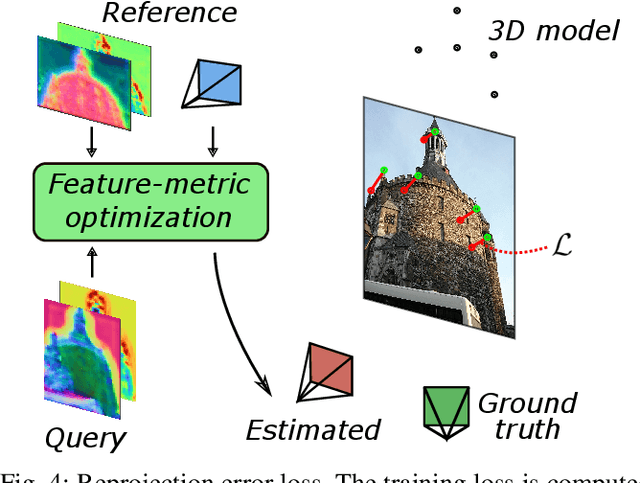
Abstract:We propose a compact pipeline to unify all the steps of Visual Localization: image retrieval, candidate re-ranking and initial pose estimation, and camera pose refinement. Our key assumption is that the deep features used for these individual tasks share common characteristics, so we should reuse them in all the procedures of the pipeline. Our DRAN (Deep Retrieval and image Alignment Network) is able to extract global descriptors for efficient image retrieval, use intermediate hierarchical features to re-rank the retrieval list and produce an intial pose guess, which is finally refined by means of a feature-metric optimization based on learned deep multi-scale dense features. DRAN is the first single network able to produce the features for the three steps of visual localization. DRAN achieves a competitive performance in terms of robustness and accuracy specially in extreme day-night changes.
SD-DefSLAM: Semi-Direct Monocular SLAM for Deformable and Intracorporeal Scenes
Oct 19, 2020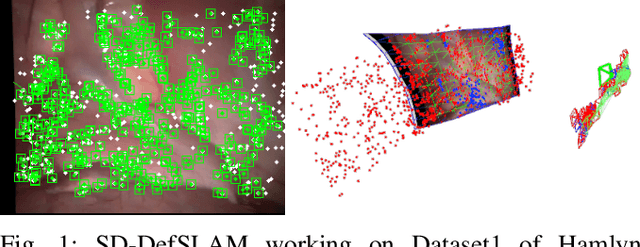
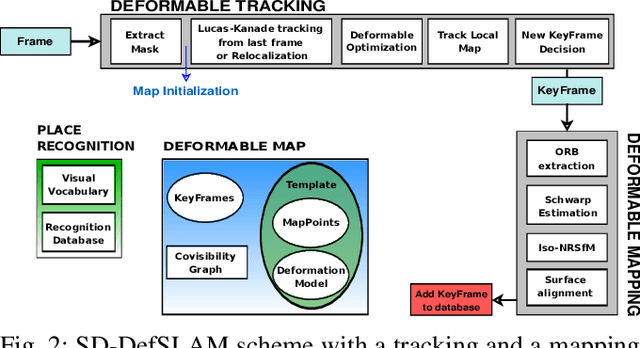
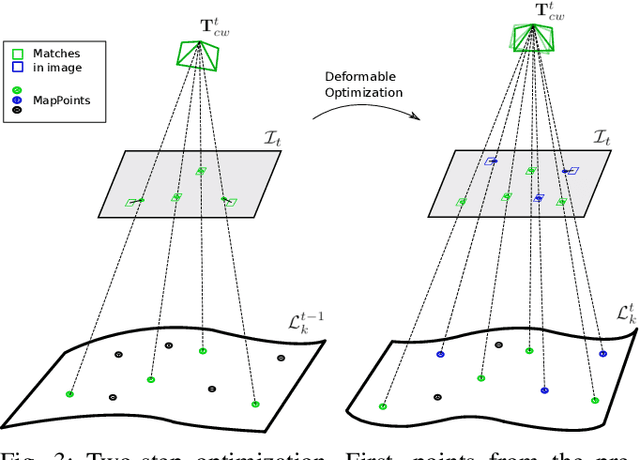
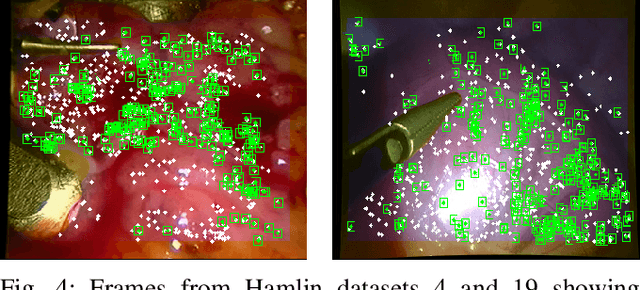
Abstract:Conventional SLAM techniques strongly rely on scene rigidity to solve data association, ignoring dynamic parts of the scene. In this work we present Semi-Direct DefSLAM (SD-DefSLAM), a novel monocular deformable SLAM method able to map highly deforming environments, built on top of DefSLAM. To robustly solve data association in challenging deforming scenes, SD-DefSLAM combines direct and indirect methods: an enhanced illumination-invariant Lucas-Kanade tracker for data association, geometric Bundle Adjustment for pose and deformable map estimation, and bag-of-words based on feature descriptors for camera relocation. Dynamic objects are detected and segmented-out using a CNN trained for the specific application domain. We thoroughly evaluate our system in two public datasets. The mandala dataset is a SLAM benchmark with increasingly aggressive deformations. The Hamlyn dataset contains intracorporeal sequences that pose serious real-life challenges beyond deformation like weak texture, specular reflections, surgical tools and occlusions. Our results show that SD-DefSLAM outperforms DefSLAM in point tracking, reconstruction accuracy and scale drift thanks to the improvement in all the data association steps, being the first system able to robustly perform SLAM inside the human body.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge