Baoxin Li
FID-Net: A Feature-Enhanced Deep Learning Network for Forest Infestation Detection
Dec 15, 2025Abstract:Forest pests threaten ecosystem stability, requiring efficient monitoring. To overcome the limitations of traditional methods in large-scale, fine-grained detection, this study focuses on accurately identifying infected trees and analyzing infestation patterns. We propose FID-Net, a deep learning model that detects pest-affected trees from UAV visible-light imagery and enables infestation analysis via three spatial metrics. Based on YOLOv8n, FID-Net introduces a lightweight Feature Enhancement Module (FEM) to extract disease-sensitive cues, an Adaptive Multi-scale Feature Fusion Module (AMFM) to align and fuse dual-branch features (RGB and FEM-enhanced), and an Efficient Channel Attention (ECA) mechanism to enhance discriminative information efficiently. From detection results, we construct a pest situation analysis framework using: (1) Kernel Density Estimation to locate infection hotspots; (2) neighborhood evaluation to assess healthy trees' infection risk; (3) DBSCAN clustering to identify high-density healthy clusters as priority protection zones. Experiments on UAV imagery from 32 forest plots in eastern Tianshan, China, show that FID-Net achieves 86.10% precision, 75.44% recall, 82.29% mAP@0.5, and 64.30% mAP@0.5:0.95, outperforming mainstream YOLO models. Analysis confirms infected trees exhibit clear clustering, supporting targeted forest protection. FID-Net enables accurate tree health discrimination and, combined with spatial metrics, provides reliable data for intelligent pest monitoring, early warning, and precise management.
Context-Aware Token Selection and Packing for Enhanced Vision Transformer
Oct 31, 2024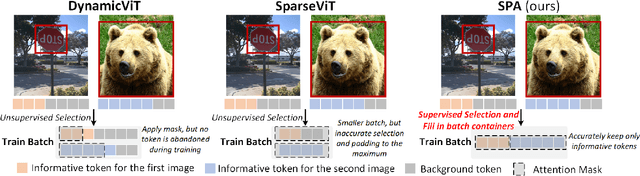

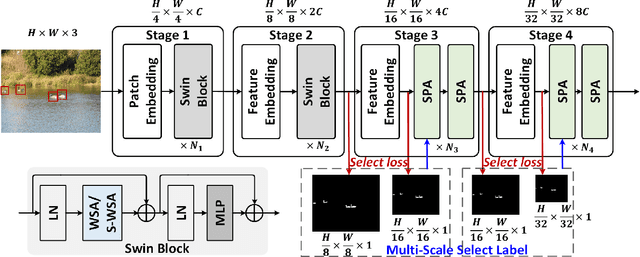
Abstract:In recent years, the long-range attention mechanism of vision transformers has driven significant performance breakthroughs across various computer vision tasks. However, the traditional self-attention mechanism, which processes both informative and non-informative tokens, suffers from inefficiency and inaccuracies. While sparse attention mechanisms have been introduced to mitigate these issues by pruning tokens involved in attention, they often lack context-awareness and intelligence. These mechanisms frequently apply a uniform token selection strategy across different inputs for batch training or optimize efficiency only for the inference stage. To overcome these challenges, we propose a novel algorithm: Select and Pack Attention (SPA). SPA dynamically selects informative tokens using a low-cost gating layer supervised by selection labels and packs these tokens into new batches, enabling a variable number of tokens to be used in parallelized GPU batch training and inference. Extensive experiments across diverse datasets and computer vision tasks demonstrate that SPA delivers superior performance and efficiency, including a 0.6 mAP improvement in object detection and a 16.4% reduction in computational costs.
Domain Adaptation Using Pseudo Labels
Feb 09, 2024Abstract:In the absence of labeled target data, unsupervised domain adaptation approaches seek to align the marginal distributions of the source and target domains in order to train a classifier for the target. Unsupervised domain alignment procedures are category-agnostic and end up misaligning the categories. We address this problem by deploying a pretrained network to determine accurate labels for the target domain using a multi-stage pseudo-label refinement procedure. The filters are based on the confidence, distance (conformity), and consistency of the pseudo labels. Our results on multiple datasets demonstrate the effectiveness of our simple procedure in comparison with complex state-of-the-art techniques.
Transformer-based Selective Super-Resolution for Efficient Image Refinement
Dec 10, 2023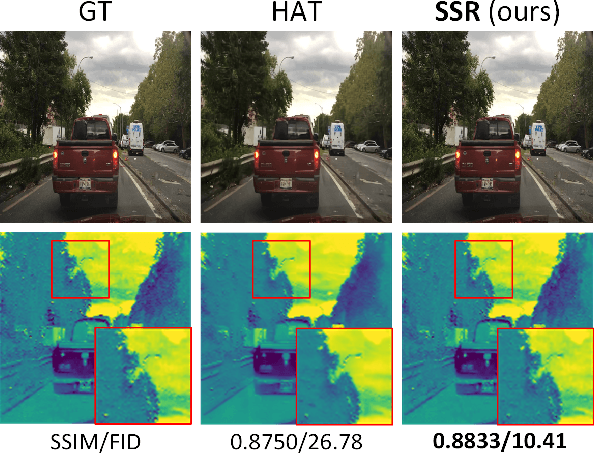
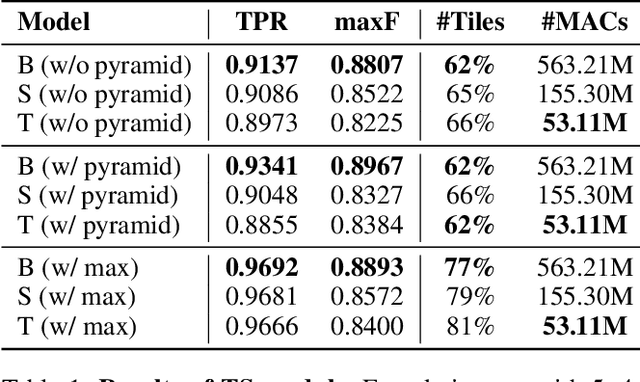
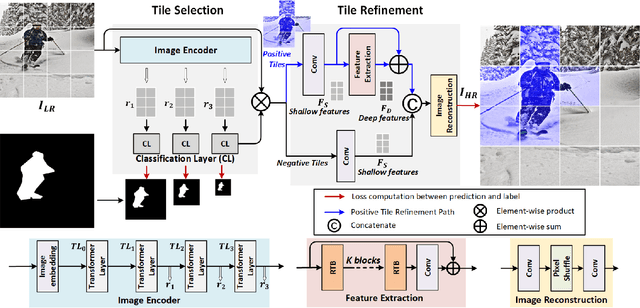
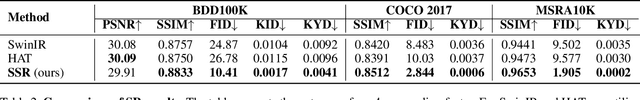
Abstract:Conventional super-resolution methods suffer from two drawbacks: substantial computational cost in upscaling an entire large image, and the introduction of extraneous or potentially detrimental information for downstream computer vision tasks during the refinement of the background. To solve these issues, we propose a novel transformer-based algorithm, Selective Super-Resolution (SSR), which partitions images into non-overlapping tiles, selects tiles of interest at various scales with a pyramid architecture, and exclusively reconstructs these selected tiles with deep features. Experimental results on three datasets demonstrate the efficiency and robust performance of our approach for super-resolution. Compared to the state-of-the-art methods, the FID score is reduced from 26.78 to 10.41 with 40% reduction in computation cost for the BDD100K dataset. The source code is available at https://github.com/destiny301/SSR.
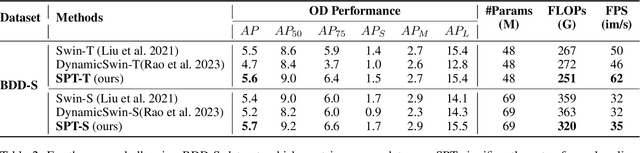
Patch-based Selection and Refinement for Early Object Detection
Nov 03, 2023



Abstract:Early object detection (OD) is a crucial task for the safety of many dynamic systems. Current OD algorithms have limited success for small objects at a long distance. To improve the accuracy and efficiency of such a task, we propose a novel set of algorithms that divide the image into patches, select patches with objects at various scales, elaborate the details of a small object, and detect it as early as possible. Our approach is built upon a transformer-based network and integrates the diffusion model to improve the detection accuracy. As demonstrated on BDD100K, our algorithms enhance the mAP for small objects from 1.03 to 8.93, and reduce the data volume in computation by more than 77\%. The source code is available at \href{https://github.com/destiny301/dpr}{https://github.com/destiny301/dpr}
Instance Adaptive Prototypical Contrastive Embedding for Generalized Zero Shot Learning
Sep 14, 2023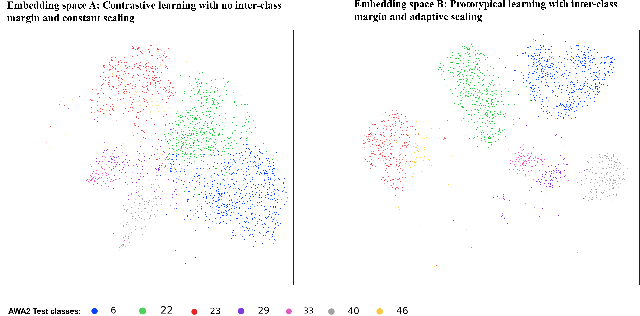
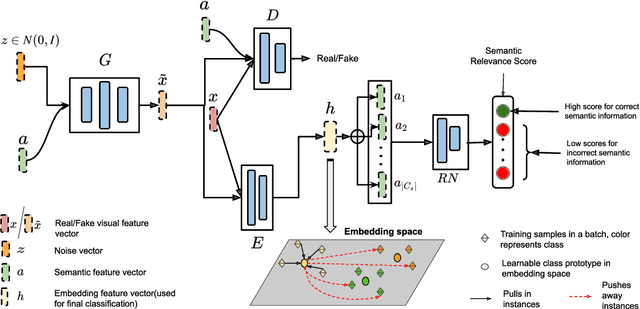
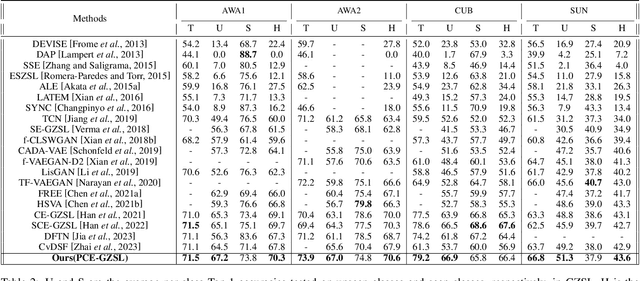
Abstract:Generalized zero-shot learning(GZSL) aims to classify samples from seen and unseen labels, assuming unseen labels are not accessible during training. Recent advancements in GZSL have been expedited by incorporating contrastive-learning-based (instance-based) embedding in generative networks and leveraging the semantic relationship between data points. However, existing embedding architectures suffer from two limitations: (1) limited discriminability of synthetic features' embedding without considering fine-grained cluster structures; (2) inflexible optimization due to restricted scaling mechanisms on existing contrastive embedding networks, leading to overlapped representations in the embedding space. To enhance the quality of representations in the embedding space, as mentioned in (1), we propose a margin-based prototypical contrastive learning embedding network that reaps the benefits of prototype-data (cluster quality enhancement) and implicit data-data (fine-grained representations) interaction while providing substantial cluster supervision to the embedding network and the generator. To tackle (2), we propose an instance adaptive contrastive loss that leads to generalized representations for unseen labels with increased inter-class margin. Through comprehensive experimental evaluation, we show that our method can outperform the current state-of-the-art on three benchmark datasets. Our approach also consistently achieves the best unseen performance in the GZSL setting.
Deep-Learning-based Fast and Accurate 3D CT Deformable Image Registration in Lung Cancer
Apr 21, 2023Abstract:Purpose: In some proton therapy facilities, patient alignment relies on two 2D orthogonal kV images, taken at fixed, oblique angles, as no 3D on-the-bed imaging is available. The visibility of the tumor in kV images is limited since the patient's 3D anatomy is projected onto a 2D plane, especially when the tumor is behind high-density structures such as bones. This can lead to large patient setup errors. A solution is to reconstruct the 3D CT image from the kV images obtained at the treatment isocenter in the treatment position. Methods: An asymmetric autoencoder-like network built with vision-transformer blocks was developed. The data was collected from 1 head and neck patient: 2 orthogonal kV images (1024x1024 voxels), 1 3D CT with padding (512x512x512) acquired from the in-room CT-on-rails before kVs were taken and 2 digitally-reconstructed-radiograph (DRR) images (512x512) based on the CT. We resampled kV images every 8 voxels and DRR and CT every 4 voxels, thus formed a dataset consisting of 262,144 samples, in which the images have a dimension of 128 for each direction. In training, both kV and DRR images were utilized, and the encoder was encouraged to learn the jointed feature map from both kV and DRR images. In testing, only independent kV images were used. The full-size synthetic CT (sCT) was achieved by concatenating the sCTs generated by the model according to their spatial information. The image quality of the synthetic CT (sCT) was evaluated using mean absolute error (MAE) and per-voxel-absolute-CT-number-difference volume histogram (CDVH). Results: The model achieved a speed of 2.1s and a MAE of <40HU. The CDVH showed that <5% of the voxels had a per-voxel-absolute-CT-number-difference larger than 185 HU. Conclusion: A patient-specific vision-transformer-based network was developed and shown to be accurate and efficient to reconstruct 3D CT images from kV images.
Brainomaly: Unsupervised Neurologic Disease Detection Utilizing Unannotated T1-weighted Brain MR Images
Feb 18, 2023Abstract:Deep neural networks have revolutionized the field of supervised learning by enabling accurate predictions through learning from large annotated datasets. However, acquiring large annotated medical imaging datasets is a challenging task, especially for rare diseases, due to the high cost, time, and effort required for annotation. In these scenarios, unsupervised disease detection methods, such as anomaly detection, can save significant human effort. A typically used approach for anomaly detection is to learn the images from healthy subjects only, assuming the model will detect the images from diseased subjects as outliers. However, in many real-world scenarios, unannotated datasets with a mix of healthy and diseased individuals are available. Recent studies have shown improvement in unsupervised disease/anomaly detection using such datasets of unannotated images from healthy and diseased individuals compared to datasets that only include images from healthy individuals. A major issue remains unaddressed in these studies, which is selecting the best model for inference from a set of trained models without annotated samples. To address this issue, we propose Brainomaly, a GAN-based image-to-image translation method for neurologic disease detection using unannotated T1-weighted brain MRIs of individuals with neurologic diseases and healthy subjects. Brainomaly is trained to remove the diseased regions from the input brain MRIs and generate MRIs of corresponding healthy brains. Instead of generating the healthy images directly, Brainomaly generates an additive map where each voxel indicates the amount of changes required to make the input image look healthy. In addition, Brainomaly uses a pseudo-AUC metric for inference model selection, which further improves the detection performance. Our Brainomaly outperforms existing state-of-the-art methods by large margins.
PatchRot: A Self-Supervised Technique for Training Vision Transformers
Oct 27, 2022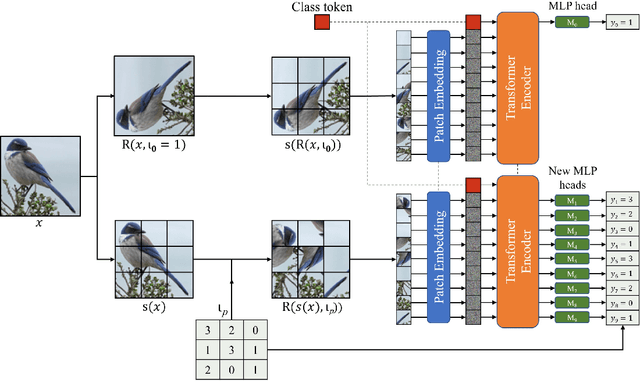
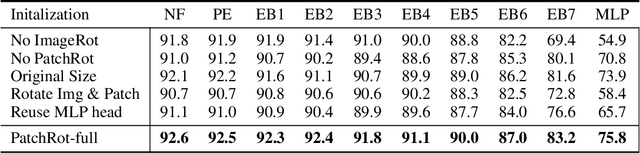
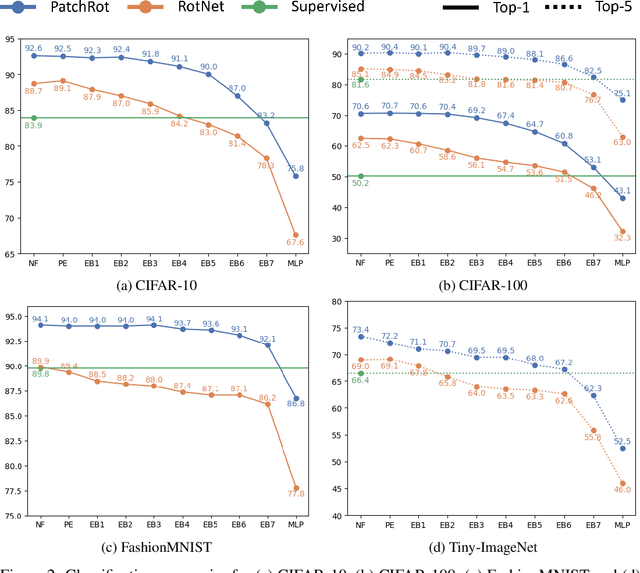
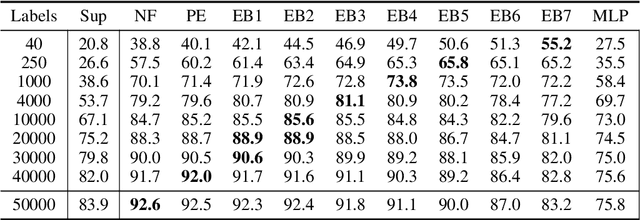
Abstract:Vision transformers require a huge amount of labeled data to outperform convolutional neural networks. However, labeling a huge dataset is a very expensive process. Self-supervised learning techniques alleviate this problem by learning features similar to supervised learning in an unsupervised way. In this paper, we propose a self-supervised technique PatchRot that is crafted for vision transformers. PatchRot rotates images and image patches and trains the network to predict the rotation angles. The network learns to extract both global and local features from an image. Our extensive experiments on different datasets showcase PatchRot training learns rich features which outperform supervised learning and compared baseline.
HealthyGAN: Learning from Unannotated Medical Images to Detect Anomalies Associated with Human Disease
Sep 05, 2022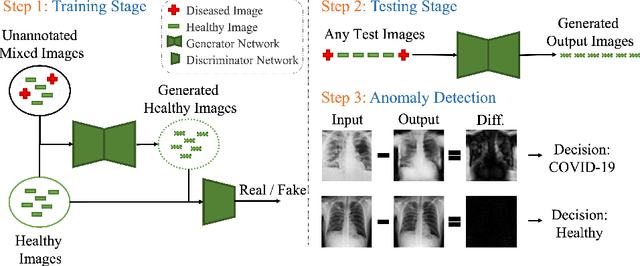
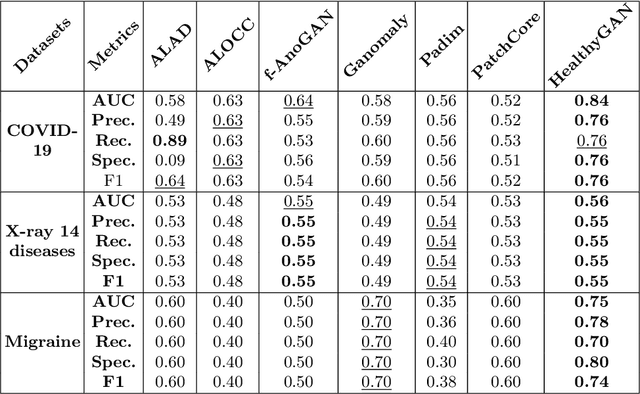
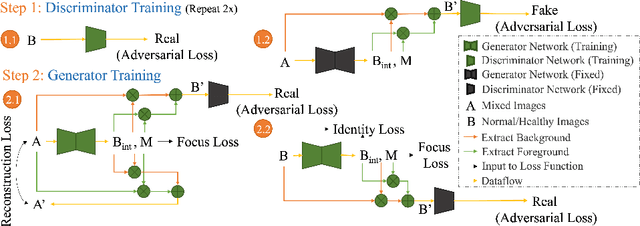
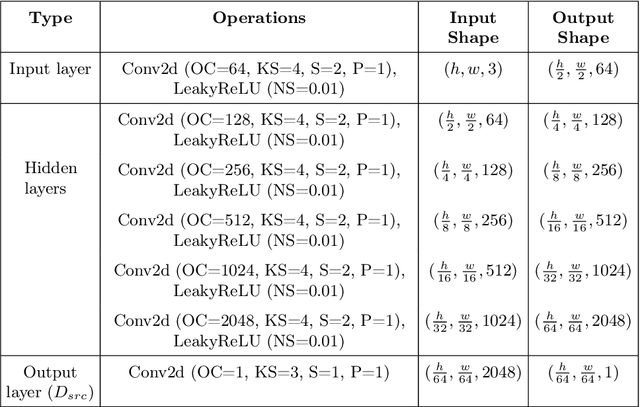
Abstract:Automated anomaly detection from medical images, such as MRIs and X-rays, can significantly reduce human effort in disease diagnosis. Owing to the complexity of modeling anomalies and the high cost of manual annotation by domain experts (e.g., radiologists), a typical technique in the current medical imaging literature has focused on deriving diagnostic models from healthy subjects only, assuming the model will detect the images from patients as outliers. However, in many real-world scenarios, unannotated datasets with a mix of both healthy and diseased individuals are abundant. Therefore, this paper poses the research question of how to improve unsupervised anomaly detection by utilizing (1) an unannotated set of mixed images, in addition to (2) the set of healthy images as being used in the literature. To answer the question, we propose HealthyGAN, a novel one-directional image-to-image translation method, which learns to translate the images from the mixed dataset to only healthy images. Being one-directional, HealthyGAN relaxes the requirement of cycle consistency of existing unpaired image-to-image translation methods, which is unattainable with mixed unannotated data. Once the translation is learned, we generate a difference map for any given image by subtracting its translated output. Regions of significant responses in the difference map correspond to potential anomalies (if any). Our HealthyGAN outperforms the conventional state-of-the-art methods by significant margins on two publicly available datasets: COVID-19 and NIH ChestX-ray14, and one institutional dataset collected from Mayo Clinic. The implementation is publicly available at https://github.com/mahfuzmohammad/HealthyGAN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge