Jay Shah
DinoAtten3D: Slice-Level Attention Aggregation of DinoV2 for 3D Brain MRI Anomaly Classification
Sep 15, 2025Abstract:Anomaly detection and classification in medical imaging are critical for early diagnosis but remain challenging due to limited annotated data, class imbalance, and the high cost of expert labeling. Emerging vision foundation models such as DINOv2, pretrained on extensive, unlabeled datasets, offer generalized representations that can potentially alleviate these limitations. In this study, we propose an attention-based global aggregation framework tailored specifically for 3D medical image anomaly classification. Leveraging the self-supervised DINOv2 model as a pretrained feature extractor, our method processes individual 2D axial slices of brain MRIs, assigning adaptive slice-level importance weights through a soft attention mechanism. To further address data scarcity, we employ a composite loss function combining supervised contrastive learning with class-variance regularization, enhancing inter-class separability and intra-class consistency. We validate our framework on the ADNI dataset and an institutional multi-class headache cohort, demonstrating strong anomaly classification performance despite limited data availability and significant class imbalance. Our results highlight the efficacy of utilizing pretrained 2D foundation models combined with attention-based slice aggregation for robust volumetric anomaly detection in medical imaging. Our implementation is publicly available at https://github.com/Rafsani/DinoAtten3D.git.
CREASE-2D Analysis of Small Angle X-ray Scattering Data from Supramolecular Dipeptide Systems
Apr 04, 2025



Abstract:In this paper, we extend a recently developed machine-learning (ML) based CREASE-2D method to analyze the entire two-dimensional (2D) scattering pattern obtained from small angle X-ray scattering measurements of supramolecular dipeptide micellar systems. Traditional analysis of such scattering data would involve use of approximate or incorrect analytical models to fit to azimuthally-averaged 1D scattering patterns that can miss the anisotropic arrangements. Analysis of the 2D scattering profiles of such micellar solutions using CREASE-2D allows us to understand both isotropic and anisotropic structural arrangements that are present in these systems of assembled dipeptides in water and in the presence of added solvents/salts. CREASE-2D outputs distributions of relevant structural features including ones that cannot be identified with existing analytical models (e.g., assembled tubes, cross-sectional eccentricity, tortuosity, orientational order). The representative three-dimensional (3D) real-space structures for the optimized values of these structural features further facilitate visualization of the structures. Through this detailed interpretation of these 2D SAXS profiles we are able to characterize the shapes of the assembled tube structures as a function of dipeptide chemistry, solution conditions with varying salts and solvents, and relative concentrations of all components. This paper demonstrates how CREASE-2D analysis of entire SAXS profiles can provide an unprecedented level of understanding of structural arrangements which has not been possible through traditional analytical model fits to the 1D SAXS data.
FlashAttention-3: Fast and Accurate Attention with Asynchrony and Low-precision
Jul 11, 2024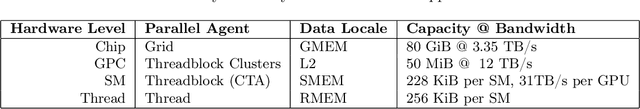
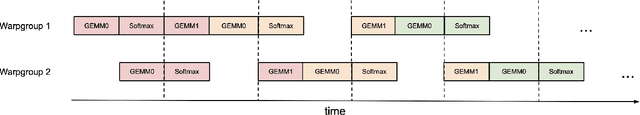
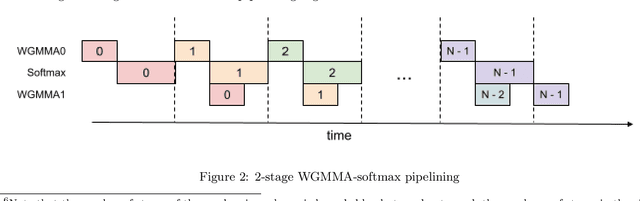

Abstract:Attention, as a core layer of the ubiquitous Transformer architecture, is the bottleneck for large language models and long-context applications. FlashAttention elaborated an approach to speed up attention on GPUs through minimizing memory reads/writes. However, it has yet to take advantage of new capabilities present in recent hardware, with FlashAttention-2 achieving only 35% utilization on the H100 GPU. We develop three main techniques to speed up attention on Hopper GPUs: exploiting asynchrony of the Tensor Cores and TMA to (1) overlap overall computation and data movement via warp-specialization and (2) interleave block-wise matmul and softmax operations, and (3) block quantization and incoherent processing that leverages hardware support for FP8 low-precision. We demonstrate that our method, FlashAttention-3, achieves speedup on H100 GPUs by 1.5-2.0$\times$ with FP16 reaching up to 740 TFLOPs/s (75% utilization), and with FP8 reaching close to 1.2 PFLOPs/s. We validate that FP8 FlashAttention-3 achieves 2.6$\times$ lower numerical error than a baseline FP8 attention.
AnoFPDM: Anomaly Segmentation with Forward Process of Diffusion Models for Brain MRI
Apr 24, 2024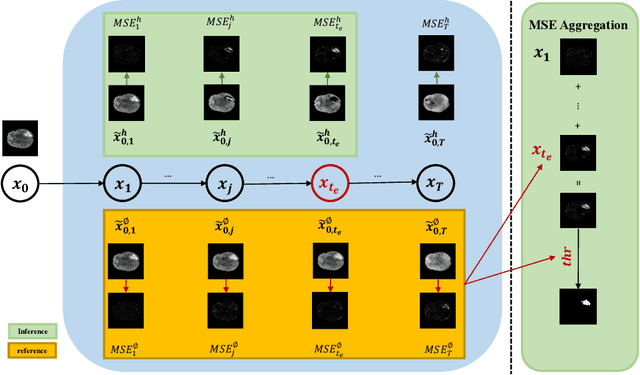

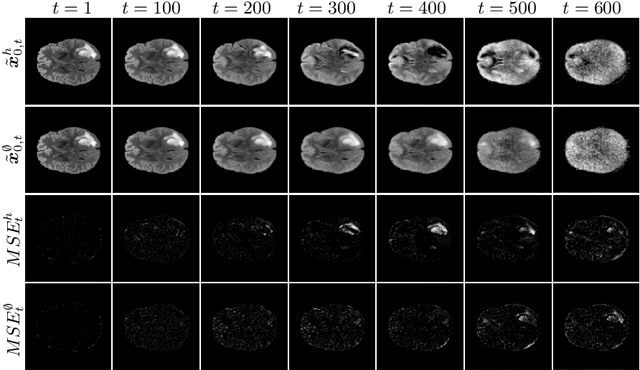

Abstract:Weakly-supervised diffusion models (DM) in anomaly segmentation, leveraging image-level labels, have attracted significant attention for their superior performance compared to unsupervised methods. It eliminates the need for pixel-level labels in training, offering a more cost-effective alternative to supervised methods. However, existing methods are not fully weakly-supervised because they heavily rely on costly pixel-level labels for hyperparameter tuning in inference. To tackle this challenge, we introduce Anomaly Segmentation with Forward Process of Diffusion Models (AnoFPDM), a fully weakly-supervised framework that operates without the need for pixel-level labels. Leveraging the unguided forward process as a reference, we identify suitable hyperparameters, i.e., noise scale and threshold, for each input image. We aggregate anomaly maps from each step in the forward process, enhancing the signal strength of anomalous regions. Remarkably, our proposed method outperforms recent state-of-the-art weakly-supervised approaches, even without utilizing pixel-level labels.
A Case Study in CUDA Kernel Fusion: Implementing FlashAttention-2 on NVIDIA Hopper Architecture using the CUTLASS Library
Dec 19, 2023Abstract:We provide an optimized implementation of the forward pass of FlashAttention-2, a popular memory-aware scaled dot-product attention algorithm, as a custom fused CUDA kernel targeting NVIDIA Hopper architecture and written using the open-source CUTLASS library. In doing so, we explain the challenges and techniques involved in fusing online-softmax with back-to-back GEMM kernels, utilizing the Hopper-specific Tensor Memory Accelerator (TMA) and Warpgroup Matrix-Multiply-Accumulate (WGMMA) instructions, defining and transforming CUTLASS Layouts and Tensors, overlapping copy and GEMM operations, and choosing optimal tile sizes for the Q, K and V attention matrices while balancing the register pressure and shared memory utilization. In head-to-head benchmarks on a single H100 PCIe GPU for some common choices of hyperparameters, we observe 20-50% higher FLOPs/s over a version of FlashAttention-2 optimized for last-generation NVIDIA Ampere architecture.
Body-mounted MR-conditional Robot for Minimally Invasive Liver Intervention
Oct 11, 2023Abstract:MR-guided microwave ablation (MWA) has proven effective in treating hepatocellular carcinoma (HCC) with small-sized tumors, but the state-of-the-art technique suffers from sub-optimal workflow due to speed and accuracy of needle placement. This paper presents a compact body-mounted MR-conditional robot that can operate in closed-bore MR scanners for accurate needle guidance. The robotic platform consists of two stacked Cartesian XY stages, each with two degrees of freedom, that facilitate needle guidance. The robot is actuated using 3D-printed pneumatic turbines with MR-conditional bevel gear transmission systems. Pneumatic valves and control mechatronics are located inside the MRI control room and are connected to the robot with pneumatic transmission lines and optical fibers. Free space experiments indicated robot-assisted needle insertion error of 2.6$\pm$1.3 mm at an insertion depth of 80 mm. The MR-guided phantom studies were conducted to verify the MR-conditionality and targeting performance of the robot. Future work will focus on the system optimization and validations in animal trials.
Brainomaly: Unsupervised Neurologic Disease Detection Utilizing Unannotated T1-weighted Brain MR Images
Feb 18, 2023Abstract:Deep neural networks have revolutionized the field of supervised learning by enabling accurate predictions through learning from large annotated datasets. However, acquiring large annotated medical imaging datasets is a challenging task, especially for rare diseases, due to the high cost, time, and effort required for annotation. In these scenarios, unsupervised disease detection methods, such as anomaly detection, can save significant human effort. A typically used approach for anomaly detection is to learn the images from healthy subjects only, assuming the model will detect the images from diseased subjects as outliers. However, in many real-world scenarios, unannotated datasets with a mix of healthy and diseased individuals are available. Recent studies have shown improvement in unsupervised disease/anomaly detection using such datasets of unannotated images from healthy and diseased individuals compared to datasets that only include images from healthy individuals. A major issue remains unaddressed in these studies, which is selecting the best model for inference from a set of trained models without annotated samples. To address this issue, we propose Brainomaly, a GAN-based image-to-image translation method for neurologic disease detection using unannotated T1-weighted brain MRIs of individuals with neurologic diseases and healthy subjects. Brainomaly is trained to remove the diseased regions from the input brain MRIs and generate MRIs of corresponding healthy brains. Instead of generating the healthy images directly, Brainomaly generates an additive map where each voxel indicates the amount of changes required to make the input image look healthy. In addition, Brainomaly uses a pseudo-AUC metric for inference model selection, which further improves the detection performance. Our Brainomaly outperforms existing state-of-the-art methods by large margins.
HealthyGAN: Learning from Unannotated Medical Images to Detect Anomalies Associated with Human Disease
Sep 05, 2022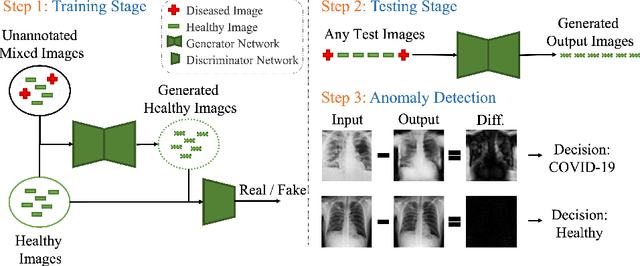
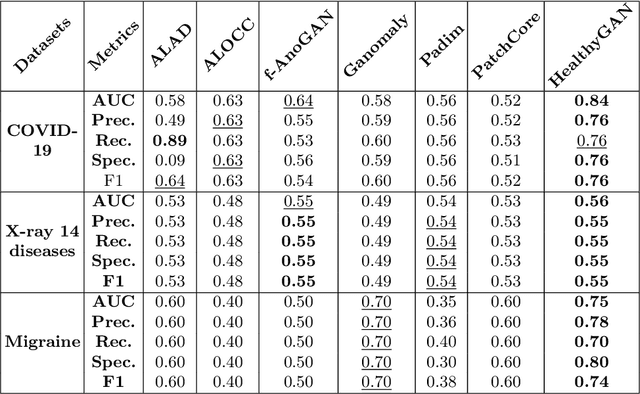
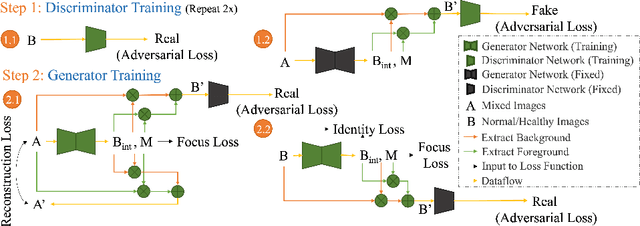
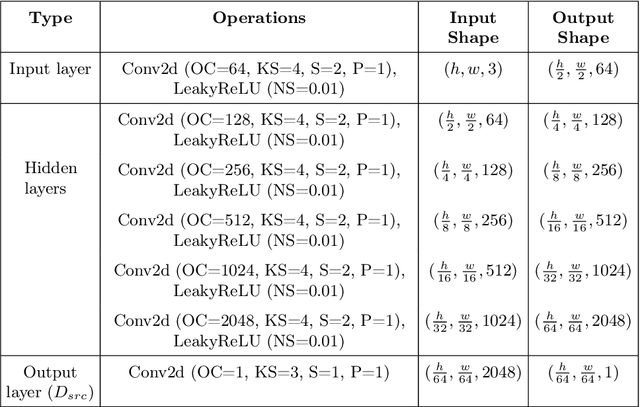
Abstract:Automated anomaly detection from medical images, such as MRIs and X-rays, can significantly reduce human effort in disease diagnosis. Owing to the complexity of modeling anomalies and the high cost of manual annotation by domain experts (e.g., radiologists), a typical technique in the current medical imaging literature has focused on deriving diagnostic models from healthy subjects only, assuming the model will detect the images from patients as outliers. However, in many real-world scenarios, unannotated datasets with a mix of both healthy and diseased individuals are abundant. Therefore, this paper poses the research question of how to improve unsupervised anomaly detection by utilizing (1) an unannotated set of mixed images, in addition to (2) the set of healthy images as being used in the literature. To answer the question, we propose HealthyGAN, a novel one-directional image-to-image translation method, which learns to translate the images from the mixed dataset to only healthy images. Being one-directional, HealthyGAN relaxes the requirement of cycle consistency of existing unpaired image-to-image translation methods, which is unattainable with mixed unannotated data. Once the translation is learned, we generate a difference map for any given image by subtracting its translated output. Regions of significant responses in the difference map correspond to potential anomalies (if any). Our HealthyGAN outperforms the conventional state-of-the-art methods by significant margins on two publicly available datasets: COVID-19 and NIH ChestX-ray14, and one institutional dataset collected from Mayo Clinic. The implementation is publicly available at https://github.com/mahfuzmohammad/HealthyGAN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge