Arthi Jayaraman
CREASE-2D Analysis of Small Angle X-ray Scattering Data from Supramolecular Dipeptide Systems
Apr 04, 2025



Abstract:In this paper, we extend a recently developed machine-learning (ML) based CREASE-2D method to analyze the entire two-dimensional (2D) scattering pattern obtained from small angle X-ray scattering measurements of supramolecular dipeptide micellar systems. Traditional analysis of such scattering data would involve use of approximate or incorrect analytical models to fit to azimuthally-averaged 1D scattering patterns that can miss the anisotropic arrangements. Analysis of the 2D scattering profiles of such micellar solutions using CREASE-2D allows us to understand both isotropic and anisotropic structural arrangements that are present in these systems of assembled dipeptides in water and in the presence of added solvents/salts. CREASE-2D outputs distributions of relevant structural features including ones that cannot be identified with existing analytical models (e.g., assembled tubes, cross-sectional eccentricity, tortuosity, orientational order). The representative three-dimensional (3D) real-space structures for the optimized values of these structural features further facilitate visualization of the structures. Through this detailed interpretation of these 2D SAXS profiles we are able to characterize the shapes of the assembled tube structures as a function of dipeptide chemistry, solution conditions with varying salts and solvents, and relative concentrations of all components. This paper demonstrates how CREASE-2D analysis of entire SAXS profiles can provide an unprecedented level of understanding of structural arrangements which has not been possible through traditional analytical model fits to the 1D SAXS data.
Machine Learning for Identifying Grain Boundaries in Scanning Electron Microscopy (SEM) Images of Nanoparticle Superlattices
Jan 07, 2025



Abstract:Nanoparticle superlattices consisting of ordered arrangements of nanoparticles exhibit unique optical, magnetic, and electronic properties arising from nanoparticle characteristics as well as their collective behaviors. Understanding how processing conditions influence the nanoscale arrangement and microstructure is critical for engineering materials with desired macroscopic properties. Microstructural features such as grain boundaries, lattice defects, and pores significantly affect these properties but are challenging to quantify using traditional manual analyses as they are labor-intensive and prone to errors. In this work, we present a machine learning workflow for automating grain segmentation in scanning electron microscopy (SEM) images of nanoparticle superlattices. This workflow integrates signal processing techniques, such as Radon transforms, with unsupervised learning methods like agglomerative hierarchical clustering to identify and segment grains without requiring manually annotated data. In the workflow we transform the raw pixel data into explainable numerical representation of superlattice orientations for clustering. Benchmarking results demonstrate the workflow's robustness against noisy images and edge cases, with a processing speed of four images per minute on standard computational hardware. This efficiency makes the workflow scalable to large datasets and makes it a valuable tool for integrating data-driven models into decision-making processes for material design and analysis. For example, one can use this workflow to quantify grain size distributions at varying processing conditions like temperature and pressure and using that knowledge adjust processing conditions to achieve desired superlattice orientations and grain sizes.
Machine Learning for Analyzing Atomic Force Microscopy (AFM) Images Generated from Polymer Blends
Sep 15, 2024



Abstract:In this paper we present a new machine learning workflow with unsupervised learning techniques to identify domains within atomic force microscopy images obtained from polymer films. The goal of the workflow is to identify the spatial location of the two types of polymer domains with little to no manual intervention and calculate the domain size distributions which in turn can help qualify the phase separated state of the material as macrophase or microphase ordered or disordered domains. We briefly review existing approaches used in other fields, computer vision and signal processing that can be applicable for the above tasks that happen frequently in the field of polymer science and engineering. We then test these approaches from computer vision and signal processing on the AFM image dataset to identify the strengths and limitations of each of these approaches for our first task. For our first domain segmentation task, we found that the workflow using discrete Fourier transform or discrete cosine transform with variance statistics as the feature works the best. The popular ResNet50 deep learning approach from computer vision field exhibited relatively poorer performance in the domain segmentation task for our AFM images as compared to the DFT and DCT based workflows. For the second task, for each of 144 input AFM images, we then used an existing porespy python package to calculate the domain size distribution from the output of that image from DFT based workflow. The information and open source codes we share in this paper can serve as a guide for researchers in the polymer and soft materials fields who need ML modeling and workflows for automated analyses of AFM images from polymer samples that may have crystalline or amorphous domains, sharp or rough interfaces between domains, or micro or macrophase separated domains.
Pair-Variational Autoencoders (PairVAE) for Linking and Cross-Reconstruction of Characterization Data from Complementary Structural Characterization Techniques
May 25, 2023



Abstract:In material research, structural characterization often requires multiple complementary techniques to obtain a holistic morphological view of the synthesized material. Depending on the availability of and accessibility of the different characterization techniques (e.g., scattering, microscopy, spectroscopy), each research facility or academic research lab may have access to high-throughput capability in one technique but face limitations (sample preparation, resolution, access time) with other techniques(s). Furthermore, one type of structural characterization data may be easier to interpret than another (e.g., microscopy images are easier to interpret than small angle scattering profiles). Thus, it is useful to have machine learning models that can be trained on paired structural characterization data from multiple techniques so that the model can generate one set of characterization data from the other. In this paper we demonstrate one such machine learning workflow, PairVAE, that works with data from Small Angle X-Ray Scattering (SAXS) that presents information about bulk morphology and images from Scanning Electron Microscopy (SEM) that presents two-dimensional local structural information of the sample. Using paired SAXS and SEM data of novel block copolymer assembled morphologies [open access data from Doerk G.S., et al. Science Advances. 2023 Jan 13;9(2): eadd3687], we train our PairVAE. After successful training, we demonstrate that the PairVAE can generate SEM images of the block copolymer morphology when it takes as input that sample's corresponding SAXS 2D pattern, and vice versa. This method can be extended to other soft materials morphologies as well and serves as a valuable tool for easy interpretation of 2D SAXS patterns as well as creating a database for other downstream calculations of structure-property relationships.
Self-supervised machine learning model for analysis of nanowire morphologies from transmission electron microscopy images
Mar 25, 2022
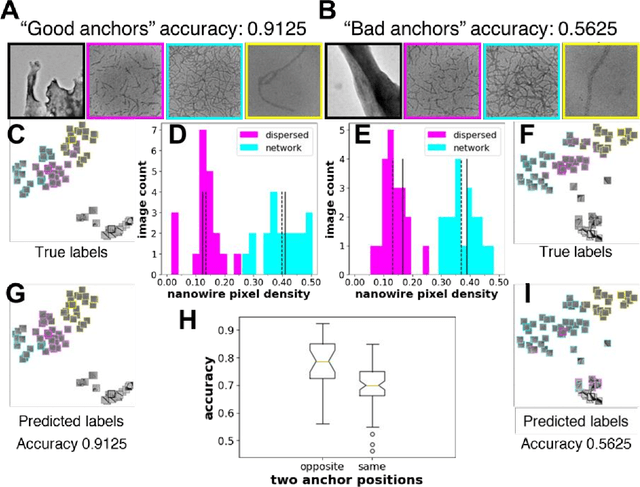
Abstract:In the field of soft materials, microscopy is the first and often only accessible method for structural characterization. There is a growing interest in the development of machine learning methods that can automate the analysis and interpretation of microscopy images. Typically training of machine learning models require large numbers of images with associated structural labels, however, manual labeling of images requires domain knowledge and is prone to human error and subjectivity. To overcome these limitations, we present a self-supervised transfer learning approach that uses a small number of labeled microscopy images for training and performs as effectively as methods trained on significantly larger data sets. Specifically, we train an image encoder with unlabeled images and use that encoder for transfer learning of different downstream image tasks (classification and segmentation) with a minimal number of labeled images for training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge