Shizhao Lu
Pair-Variational Autoencoders (PairVAE) for Linking and Cross-Reconstruction of Characterization Data from Complementary Structural Characterization Techniques
May 25, 2023



Abstract:In material research, structural characterization often requires multiple complementary techniques to obtain a holistic morphological view of the synthesized material. Depending on the availability of and accessibility of the different characterization techniques (e.g., scattering, microscopy, spectroscopy), each research facility or academic research lab may have access to high-throughput capability in one technique but face limitations (sample preparation, resolution, access time) with other techniques(s). Furthermore, one type of structural characterization data may be easier to interpret than another (e.g., microscopy images are easier to interpret than small angle scattering profiles). Thus, it is useful to have machine learning models that can be trained on paired structural characterization data from multiple techniques so that the model can generate one set of characterization data from the other. In this paper we demonstrate one such machine learning workflow, PairVAE, that works with data from Small Angle X-Ray Scattering (SAXS) that presents information about bulk morphology and images from Scanning Electron Microscopy (SEM) that presents two-dimensional local structural information of the sample. Using paired SAXS and SEM data of novel block copolymer assembled morphologies [open access data from Doerk G.S., et al. Science Advances. 2023 Jan 13;9(2): eadd3687], we train our PairVAE. After successful training, we demonstrate that the PairVAE can generate SEM images of the block copolymer morphology when it takes as input that sample's corresponding SAXS 2D pattern, and vice versa. This method can be extended to other soft materials morphologies as well and serves as a valuable tool for easy interpretation of 2D SAXS patterns as well as creating a database for other downstream calculations of structure-property relationships.
Self-supervised machine learning model for analysis of nanowire morphologies from transmission electron microscopy images
Mar 25, 2022
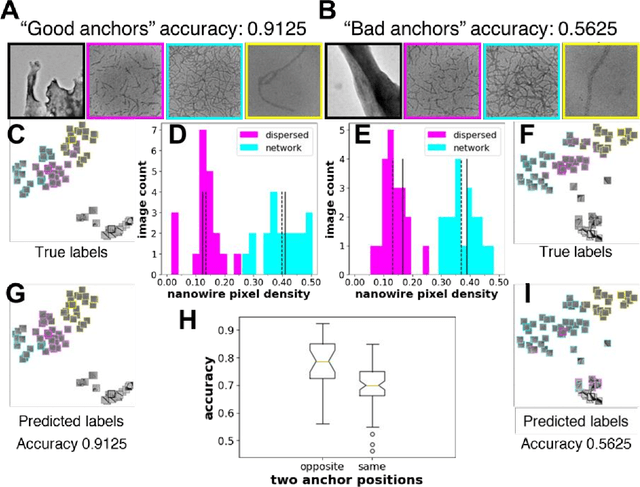
Abstract:In the field of soft materials, microscopy is the first and often only accessible method for structural characterization. There is a growing interest in the development of machine learning methods that can automate the analysis and interpretation of microscopy images. Typically training of machine learning models require large numbers of images with associated structural labels, however, manual labeling of images requires domain knowledge and is prone to human error and subjectivity. To overcome these limitations, we present a self-supervised transfer learning approach that uses a small number of labeled microscopy images for training and performs as effectively as methods trained on significantly larger data sets. Specifically, we train an image encoder with unlabeled images and use that encoder for transfer learning of different downstream image tasks (classification and segmentation) with a minimal number of labeled images for training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge