Andrew McGregor
University of Massachusetts
Improving the Efficiency of the PC Algorithm by Using Model-Based Conditional Independence Tests
Nov 12, 2022


Abstract:Learning causal structure is useful in many areas of artificial intelligence, including planning, robotics, and explanation. Constraint-based structure learning algorithms such as PC use conditional independence (CI) tests to infer causal structure. Traditionally, constraint-based algorithms perform CI tests with a preference for smaller-sized conditioning sets, partially because the statistical power of conventional CI tests declines rapidly as the size of the conditioning set increases. However, many modern conditional independence tests are model-based, and these tests use well-regularized models that maintain statistical power even with very large conditioning sets. This suggests an intriguing new strategy for constraint-based algorithms which may result in a reduction of the total number of CI tests performed: Test variable pairs with large conditioning sets first, as a pre-processing step that finds some conditional independencies quickly, before moving on to the more conventional strategy that favors small conditioning sets. We propose such a pre-processing step for the PC algorithm which relies on performing CI tests on a few randomly selected large conditioning sets. We perform an empirical analysis on directed acyclic graphs (DAGs) that correspond to real-world systems and both empirical and theoretical analyses for Erd\H{o}s-Renyi DAGs. Our results show that Pre-Processing Plus PC (P3PC) performs far fewer CI tests than the original PC algorithm, between 0.5% to 36%, and often less than 10%, of the CI tests that the PC algorithm alone performs. The efficiency gains are particularly significant for the DAGs corresponding to real-world systems.
Estimation of Entropy in Constant Space with Improved Sample Complexity
May 19, 2022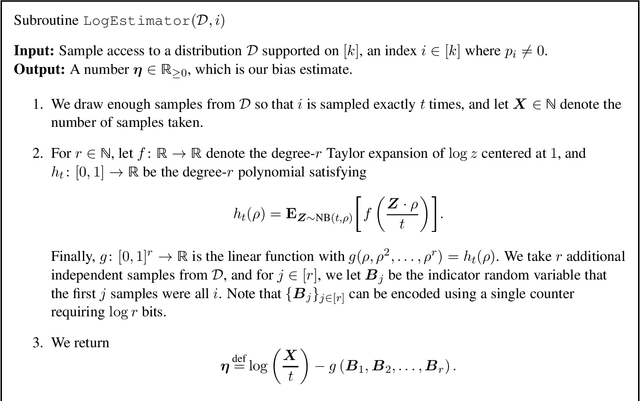
Abstract:Recent work of Acharya et al. (NeurIPS 2019) showed how to estimate the entropy of a distribution $\mathcal D$ over an alphabet of size $k$ up to $\pm\epsilon$ additive error by streaming over $(k/\epsilon^3) \cdot \text{polylog}(1/\epsilon)$ i.i.d. samples and using only $O(1)$ words of memory. In this work, we give a new constant memory scheme that reduces the sample complexity to $(k/\epsilon^2)\cdot \text{polylog}(1/\epsilon)$. We conjecture that this is optimal up to $\text{polylog}(1/\epsilon)$ factors.
Intervention Efficient Algorithms for Approximate Learning of Causal Graphs
Dec 27, 2020Abstract:We study the problem of learning the causal relationships between a set of observed variables in the presence of latents, while minimizing the cost of interventions on the observed variables. We assume access to an undirected graph $G$ on the observed variables whose edges represent either all direct causal relationships or, less restrictively, a superset of causal relationships (identified, e.g., via conditional independence tests or a domain expert). Our goal is to recover the directions of all causal or ancestral relations in $G$, via a minimum cost set of interventions. It is known that constructing an exact minimum cost intervention set for an arbitrary graph $G$ is NP-hard. We further argue that, conditioned on the hardness of approximate graph coloring, no polynomial time algorithm can achieve an approximation factor better than $\Theta(\log n)$, where $n$ is the number of observed variables in $G$. To overcome this limitation, we introduce a bi-criteria approximation goal that lets us recover the directions of all but $\epsilon n^2$ edges in $G$, for some specified error parameter $\epsilon > 0$. Under this relaxed goal, we give polynomial time algorithms that achieve intervention cost within a small constant factor of the optimal. Our algorithms combine work on efficient intervention design and the design of low-cost separating set systems, with ideas from the literature on graph property testing.
Efficient Intervention Design for Causal Discovery with Latents
May 24, 2020
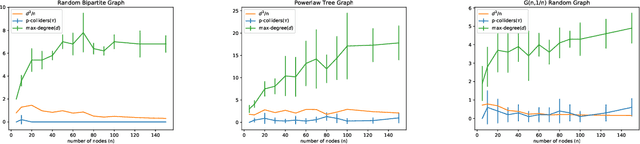
Abstract:We consider recovering a causal graph in presence of latent variables, where we seek to minimize the cost of interventions used in the recovery process. We consider two intervention cost models: (1) a linear cost model where the cost of an intervention on a subset of variables has a linear form, and (2) an identity cost model where the cost of an intervention is the same, regardless of what variables it is on, i.e., the goal is just to minimize the number of interventions. Under the linear cost model, we give an algorithm to identify the ancestral relations of the underlying causal graph, achieving within a $2$-factor of the optimal intervention cost. This approximation factor can be improved to $1+\epsilon$ for any $\epsilon > 0$ under some mild restrictions. Under the identity cost model, we bound the number of interventions needed to recover the entire causal graph, including the latent variables, using a parameterization of the causal graph through a special type of colliders. In particular, we introduce the notion of $p$-colliders, that are colliders between pair of nodes arising from a specific type of conditioning in the causal graph, and provide an upper bound on the number of interventions as a function of the maximum number of $p$-colliders between any two nodes in the causal graph.
Compact Representation of Uncertainty in Hierarchical Clustering
Feb 26, 2020



Abstract:Hierarchical clustering is a fundamental task often used to discover meaningful structures in data, such as phylogenetic trees, taxonomies of concepts, subtypes of cancer, and cascades of particle decays in particle physics. When multiple hierarchical clusterings of the data are possible, it is useful to represent uncertainty in the clustering through various probabilistic quantities. Existing approaches represent uncertainty for a range of models; however, they only provide approximate inference. This paper presents dynamic-programming algorithms and proofs for exact inference in hierarchical clustering. We are able to compute the partition function, MAP hierarchical clustering, and marginal probabilities of sub-hierarchies and clusters. Our method supports a wide range of hierarchical models and only requires a cluster compatibility function. Rather than scaling with the number of hierarchical clusterings of $n$ elements ($\omega(n n! / 2^{n-1})$), our approach runs in time and space proportional to the significantly smaller powerset of $n$. Despite still being large, these algorithms enable exact inference in small-data applications and are also interesting from a theoretical perspective. We demonstrate the utility of our method and compare its performance with respect to existing approximate methods.
Algebraic and Analytic Approaches for Parameter Learning in Mixture Models
Jan 19, 2020
Abstract:We present two different approaches for parameter learning in several mixture models in one dimension. Our first approach uses complex-analytic methods and applies to Gaussian mixtures with shared variance, binomial mixtures with shared success probability, and Poisson mixtures, among others. An example result is that $\exp(O(N^{1/3}))$ samples suffice to exactly learn a mixture of $k<N$ Poisson distributions, each with integral rate parameters bounded by $N$. Our second approach uses algebraic and combinatorial tools and applies to binomial mixtures with shared trial parameter $N$ and differing success parameters, as well as to mixtures of geometric distributions. Again, as an example, for binomial mixtures with $k$ components and success parameters discretized to resolution $\epsilon$, $O(k^2(N/\epsilon)^{8/\sqrt{\epsilon}})$ samples suffice to exactly recover the parameters. For some of these distributions, our results represent the first guarantees for parameter estimation.
Sample Complexity of Learning Mixtures of Sparse Linear Regressions
Oct 30, 2019Abstract:In the problem of learning mixtures of linear regressions, the goal is to learn a collection of signal vectors from a sequence of (possibly noisy) linear measurements, where each measurement is evaluated on an unknown signal drawn uniformly from this collection. This setting is quite expressive and has been studied both in terms of practical applications and for the sake of establishing theoretical guarantees. In this paper, we consider the case where the signal vectors are sparse; this generalizes the popular compressed sensing paradigm. We improve upon the state-of-the-art results as follows: In the noisy case, we resolve an open question of Yin et al. (IEEE Transactions on Information Theory, 2019) by showing how to handle collections of more than two vectors and present the first robust reconstruction algorithm, i.e., if the signals are not perfectly sparse, we still learn a good sparse approximation of the signals. In the noiseless case, as well as in the noisy case, we show how to circumvent the need for a restrictive assumption required in the previous work. Our techniques are quite different from those in the previous work: for the noiseless case, we rely on a property of sparse polynomials and for the noisy case, we provide new connections to learning Gaussian mixtures and use ideas from the theory of error-correcting codes.
Approximate Principal Direction Trees
Jun 18, 2012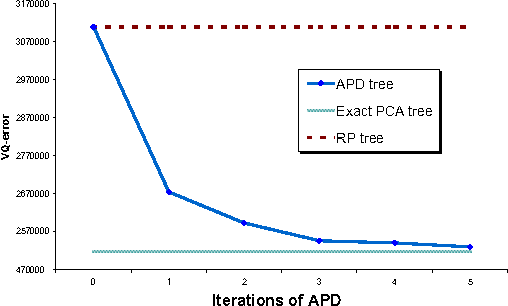

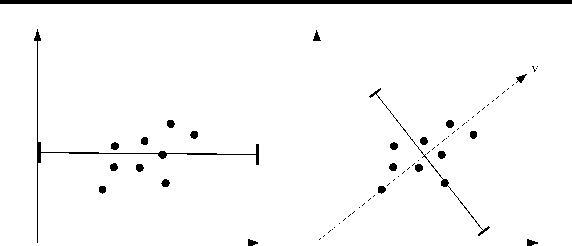
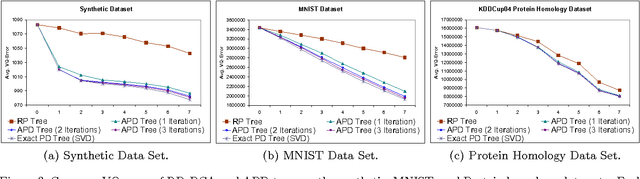
Abstract:We introduce a new spatial data structure for high dimensional data called the \emph{approximate principal direction tree} (APD tree) that adapts to the intrinsic dimension of the data. Our algorithm ensures vector-quantization accuracy similar to that of computationally-expensive PCA trees with similar time-complexity to that of lower-accuracy RP trees. APD trees use a small number of power-method iterations to find splitting planes for recursively partitioning the data. As such they provide a natural trade-off between the running-time and accuracy achieved by RP and PCA trees. Our theoretical results establish a) strong performance guarantees regardless of the convergence rate of the power-method and b) that $O(\log d)$ iterations suffice to establish the guarantee of PCA trees when the intrinsic dimension is $d$. We demonstrate this trade-off and the efficacy of our data structure on both the CPU and GPU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge