Alexandre Abraham
NEUROSPIN, PARIETAL
Improving Bias Correction Standards by Quantifying its Effects on Treatment Outcomes
Jul 20, 2024Abstract:With the growing access to administrative health databases, retrospective studies have become crucial evidence for medical treatments. Yet, non-randomized studies frequently face selection biases, requiring mitigation strategies. Propensity score matching (PSM) addresses these biases by selecting comparable populations, allowing for analysis without further methodological constraints. However, PSM has several drawbacks. Different matching methods can produce significantly different Average Treatment Effects (ATE) for the same task, even when meeting all validation criteria. To prevent cherry-picking the best method, public authorities must involve field experts and engage in extensive discussions with researchers. To address this issue, we introduce a novel metric, A2A, to reduce the number of valid matches. A2A constructs artificial matching tasks that mirror the original ones but with known outcomes, assessing each matching method's performance comprehensively from propensity estimation to ATE estimation. When combined with Standardized Mean Difference, A2A enhances the precision of model selection, resulting in a reduction of up to 50% in ATE estimation errors across synthetic tasks and up to 90% in predicted ATE variability across both synthetic and real-world datasets. To our knowledge, A2A is the first metric capable of evaluating outcome correction accuracy using covariates not involved in selection. Computing A2A requires solving hundreds of PSMs, we therefore automate all manual steps of the PSM pipeline. We integrate PSM methods from Python and R, our automated pipeline, a new metric, and reproducible experiments into popmatch, our new Python package, to enhance reproducibility and accessibility to bias correction methods.
Towards Clear Expectations for Uncertainty Estimation
Jul 27, 2022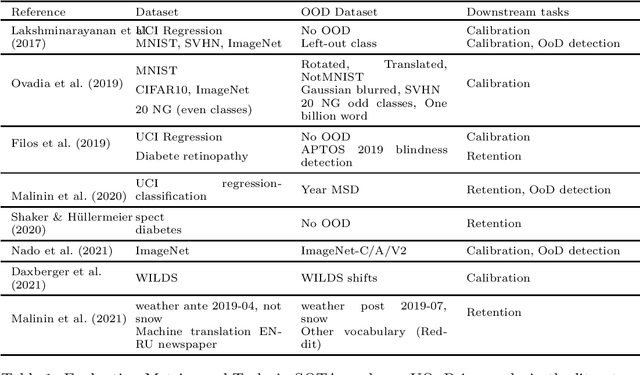
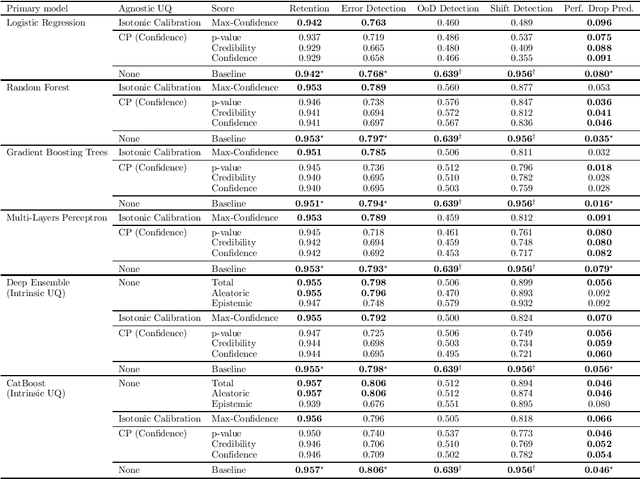
Abstract:If Uncertainty Quantification (UQ) is crucial to achieve trustworthy Machine Learning (ML), most UQ methods suffer from disparate and inconsistent evaluation protocols. We claim this inconsistency results from the unclear requirements the community expects from UQ. This opinion paper offers a new perspective by specifying those requirements through five downstream tasks where we expect uncertainty scores to have substantial predictive power. We design these downstream tasks carefully to reflect real-life usage of ML models. On an example benchmark of 7 classification datasets, we did not observe statistical superiority of state-of-the-art intrinsic UQ methods against simple baselines. We believe that our findings question the very rationale of why we quantify uncertainty and call for a standardized protocol for UQ evaluation based on metrics proven to be relevant for the ML practitioner.
Sample Noise Impact on Active Learning
Sep 03, 2021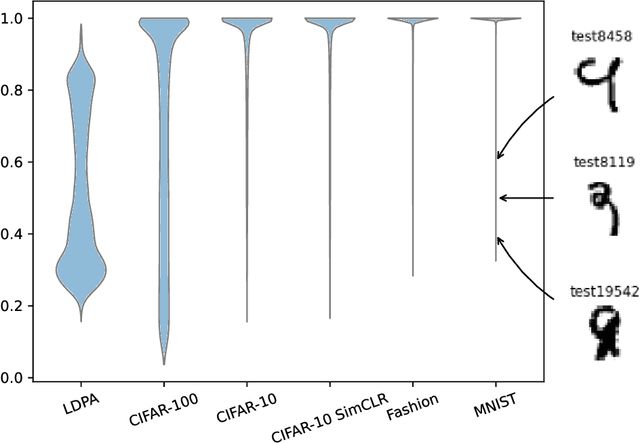
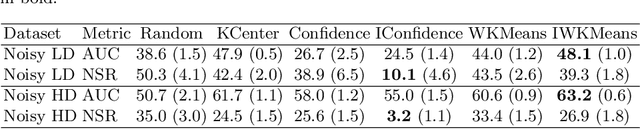
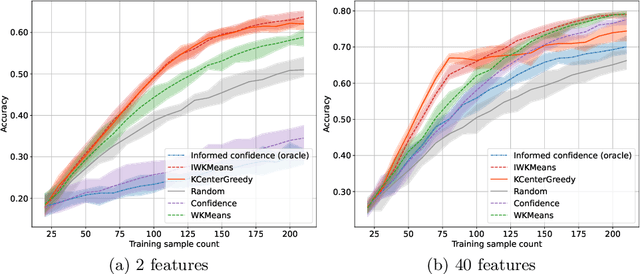
Abstract:This work explores the effect of noisy sample selection in active learning strategies. We show on both synthetic problems and real-life use-cases that knowledge of the sample noise can significantly improve the performance of active learning strategies. Building on prior work, we propose a robust sampler, Incremental Weighted K-Means that brings significant improvement on the synthetic tasks but only a marginal uplift on real-life ones. We hope that the questions raised in this paper are of interest to the community and could open new paths for active learning research.
* 9 pages, 3 figure, for the code, see https://github.com/dataiku-research/paper_ial_2021
Identification and validation of Triamcinolone and Gallopamil as treatments for early COVID-19 via an in silico repurposing pipeline
Jul 05, 2021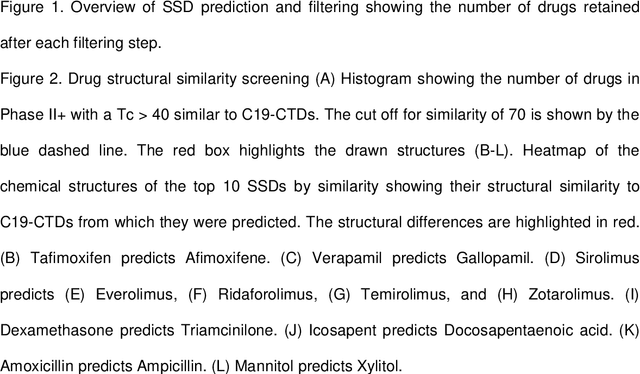
Abstract:SARS-CoV-2, the causative virus of COVID-19 continues to cause an ongoing global pandemic. Therapeutics are still needed to treat mild and severe COVID-19. Drug repurposing provides an opportunity to deploy drugs for COVID-19 more rapidly than developing novel therapeutics. Some existing drugs have shown promise for treating COVID-19 in clinical trials. This in silico study uses structural similarity to clinical trial drugs to identify two drugs with potential applications to treat early COVID-19. We apply in silico validation to suggest a possible mechanism of action for both. Triamcinolone is a corticosteroid structurally similar to Dexamethasone. Gallopamil is a calcium channel blocker structurally similar to Verapamil. We propose that both these drugs could be useful to treat early COVID-19 infection due to the proximity of their targets within a SARS-CoV-2-induced protein-protein interaction network to kinases active in early infection, and the APOA1 protein which is linked to the spread of COVID-19.
Rebuilding Trust in Active Learning with Actionable Metrics
Dec 18, 2020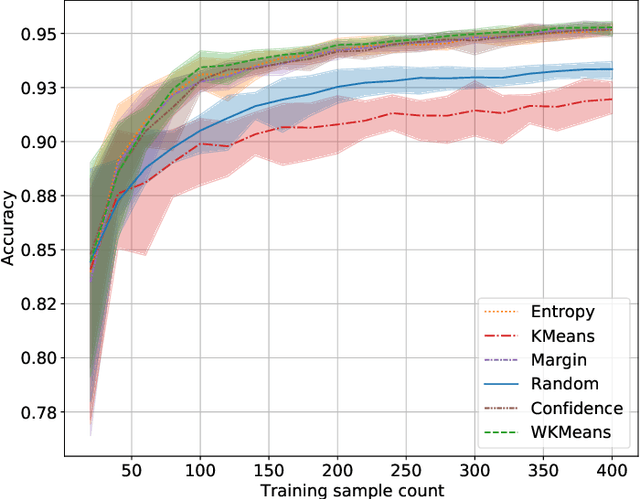
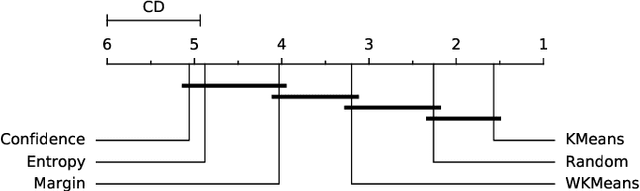
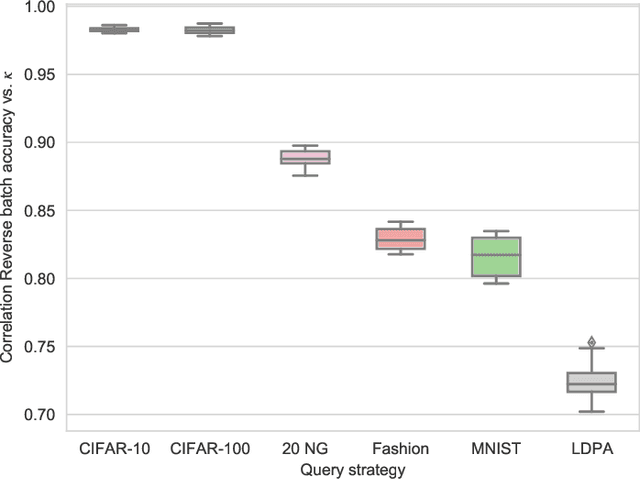
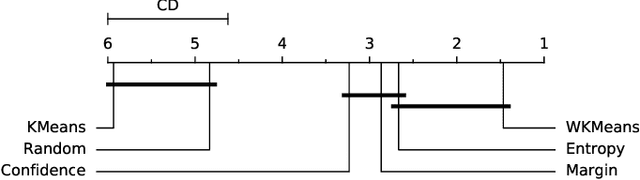
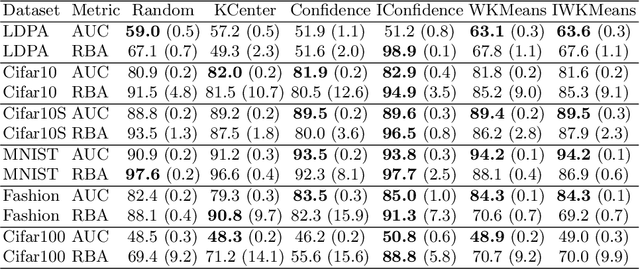
Abstract:Active Learning (AL) is an active domain of research, but is seldom used in the industry despite the pressing needs. This is in part due to a misalignment of objectives, while research strives at getting the best results on selected datasets, the industry wants guarantees that Active Learning will perform consistently and at least better than random labeling. The very one-off nature of Active Learning makes it crucial to understand how strategy selection can be carried out and what drives poor performance (lack of exploration, selection of samples that are too hard to classify, ...). To help rebuild trust of industrial practitioners in Active Learning, we present various actionable metrics. Through extensive experiments on reference datasets such as CIFAR100, Fashion-MNIST, and 20Newsgroups, we show that those metrics brings interpretability to AL strategies that can be leveraged by the practitioner.
* 16 pages, 38 figures
Offline A/B testing for Recommender Systems
Jan 22, 2018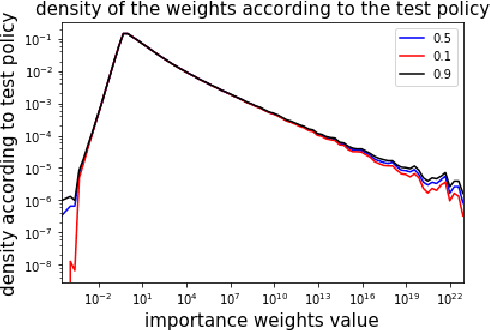

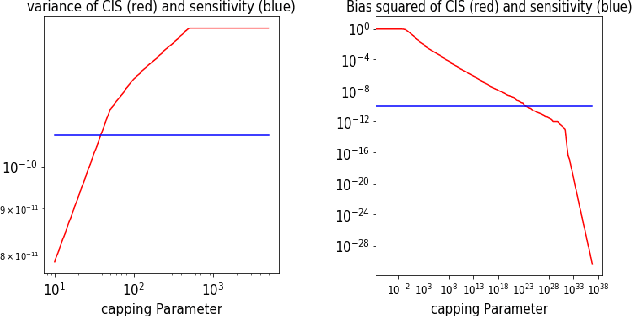
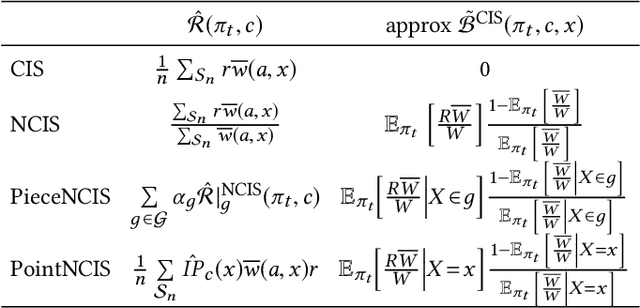
Abstract:Before A/B testing online a new version of a recommender system, it is usual to perform some offline evaluations on historical data. We focus on evaluation methods that compute an estimator of the potential uplift in revenue that could generate this new technology. It helps to iterate faster and to avoid losing money by detecting poor policies. These estimators are known as counterfactual or off-policy estimators. We show that traditional counterfactual estimators such as capped importance sampling and normalised importance sampling are experimentally not having satisfying bias-variance compromises in the context of personalised product recommendation for online advertising. We propose two variants of counterfactual estimates with different modelling of the bias that prove to be accurate in real-world conditions. We provide a benchmark of these estimators by showing their correlation with business metrics observed by running online A/B tests on a commercial recommender system.
Deriving reproducible biomarkers from multi-site resting-state data: An Autism-based example
Nov 18, 2016
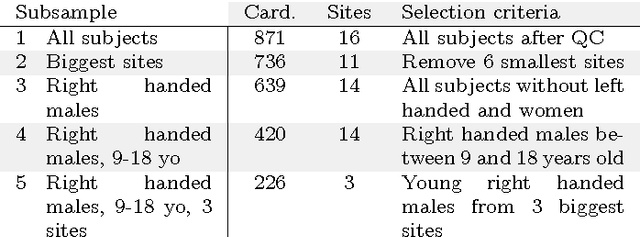
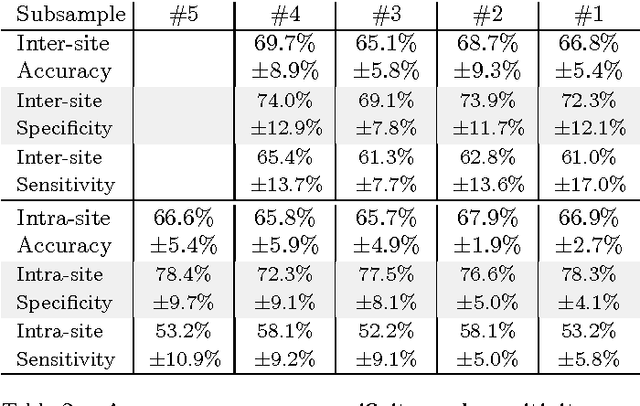

Abstract:Resting-state functional Magnetic Resonance Imaging (R-fMRI) holds the promise to reveal functional biomarkers of neuropsychiatric disorders. However, extracting such biomarkers is challenging for complex multi-faceted neuropatholo-gies, such as autism spectrum disorders. Large multi-site datasets increase sample sizes to compensate for this complexity, at the cost of uncontrolled heterogeneity. This heterogeneity raises new challenges, akin to those face in realistic diagnostic applications. Here, we demonstrate the feasibility of inter-site classification of neuropsychiatric status, with an application to the Autism Brain Imaging Data Exchange (ABIDE) database, a large (N=871) multi-site autism dataset. For this purpose, we investigate pipelines that extract the most predictive biomarkers from the data. These R-fMRI pipelines build participant-specific connectomes from functionally-defined brain areas. Connectomes are then compared across participants to learn patterns of connectivity that differentiate typical controls from individuals with autism. We predict this neuropsychiatric status for participants from the same acquisition sites or different, unseen, ones. Good choices of methods for the various steps of the pipeline lead to 67% prediction accuracy on the full ABIDE data, which is significantly better than previously reported results. We perform extensive validation on multiple subsets of the data defined by different inclusion criteria. These enables detailed analysis of the factors contributing to successful connectome-based prediction. First, prediction accuracy improves as we include more subjects, up to the maximum amount of subjects available. Second, the definition of functional brain areas is of paramount importance for biomarker discovery: brain areas extracted from large R-fMRI datasets outperform reference atlases in the classification tasks.
Region segmentation for sparse decompositions: better brain parcellations from rest fMRI
Dec 12, 2014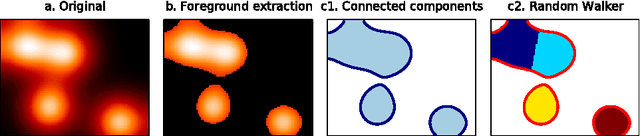
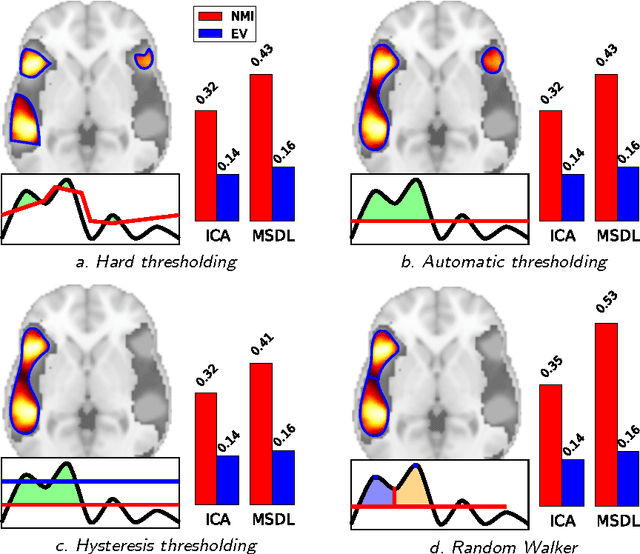
Abstract:Functional Magnetic Resonance Images acquired during resting-state provide information about the functional organization of the brain through measuring correlations between brain areas. Independent components analysis is the reference approach to estimate spatial components from weakly structured data such as brain signal time courses; each of these components may be referred to as a brain network and the whole set of components can be conceptualized as a brain functional atlas. Recently, new methods using a sparsity prior have emerged to deal with low signal-to-noise ratio data. However, even when using sophisticated priors, the results may not be very sparse and most often do not separate the spatial components into brain regions. This work presents post-processing techniques that automatically sparsify brain maps and separate regions properly using geometric operations, and compares these techniques according to faithfulness to data and stability metrics. In particular, among threshold-based approaches, hysteresis thresholding and random walker segmentation, the latter improves significantly the stability of both dense and sparse models.
Machine Learning for Neuroimaging with Scikit-Learn
Dec 12, 2014
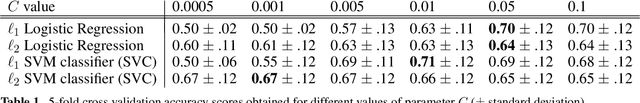
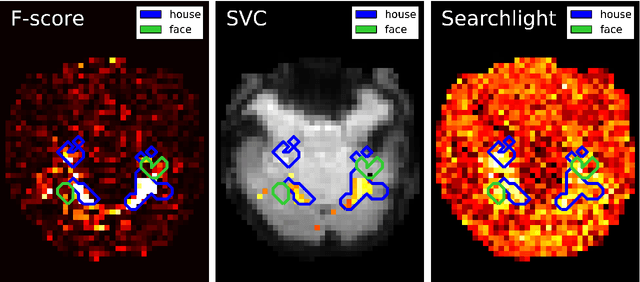

Abstract:Statistical machine learning methods are increasingly used for neuroimaging data analysis. Their main virtue is their ability to model high-dimensional datasets, e.g. multivariate analysis of activation images or resting-state time series. Supervised learning is typically used in decoding or encoding settings to relate brain images to behavioral or clinical observations, while unsupervised learning can uncover hidden structures in sets of images (e.g. resting state functional MRI) or find sub-populations in large cohorts. By considering different functional neuroimaging applications, we illustrate how scikit-learn, a Python machine learning library, can be used to perform some key analysis steps. Scikit-learn contains a very large set of statistical learning algorithms, both supervised and unsupervised, and its application to neuroimaging data provides a versatile tool to study the brain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge