Soorin Yim
Geometric Embedding Alignment via Curvature Matching in Transfer Learning
Jun 16, 2025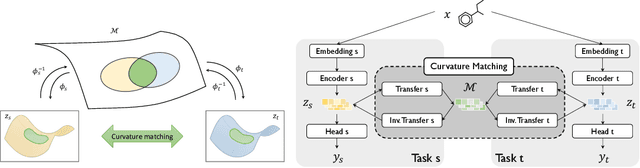

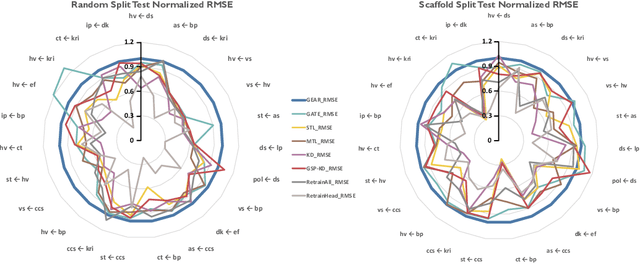
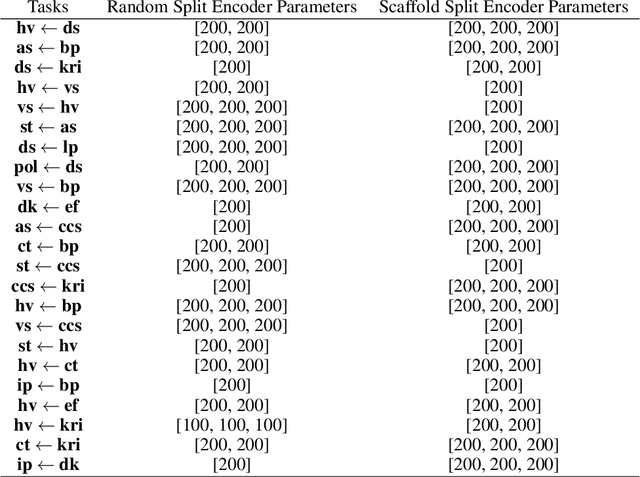
Abstract:Geometrical interpretations of deep learning models offer insightful perspectives into their underlying mathematical structures. In this work, we introduce a novel approach that leverages differential geometry, particularly concepts from Riemannian geometry, to integrate multiple models into a unified transfer learning framework. By aligning the Ricci curvature of latent space of individual models, we construct an interrelated architecture, namely Geometric Embedding Alignment via cuRvature matching in transfer learning (GEAR), which ensures comprehensive geometric representation across datapoints. This framework enables the effective aggregation of knowledge from diverse sources, thereby improving performance on target tasks. We evaluate our model on 23 molecular task pairs sourced from various domains and demonstrate significant performance gains over existing benchmark model under both random (14.4%) and scaffold (8.3%) data splits.
Scalable Multi-Task Transfer Learning for Molecular Property Prediction
Oct 01, 2024



Abstract:Molecules have a number of distinct properties whose importance and application vary. Often, in reality, labels for some properties are hard to achieve despite their practical importance. A common solution to such data scarcity is to use models of good generalization with transfer learning. This involves domain experts for designing source and target tasks whose features are shared. However, this approach has limitations: i). Difficulty in accurate design of source-target task pairs due to the large number of tasks, and ii). corresponding computational burden verifying many trials and errors of transfer learning design, thereby iii). constraining the potential of foundation modeling of multi-task molecular property prediction. We address the limitations of the manual design of transfer learning via data-driven bi-level optimization. The proposed method enables scalable multi-task transfer learning for molecular property prediction by automatically obtaining the optimal transfer ratios. Empirically, the proposed method improved the prediction performance of 40 molecular properties and accelerated training convergence.
Task Addition in Multi-Task Learning by Geometrical Alignment
Sep 25, 2024



Abstract:Training deep learning models on limited data while maintaining generalization is one of the fundamental challenges in molecular property prediction. One effective solution is transferring knowledge extracted from abundant datasets to those with scarce data. Recently, a novel algorithm called Geometrically Aligned Transfer Encoder (GATE) has been introduced, which uses soft parameter sharing by aligning the geometrical shapes of task-specific latent spaces. However, GATE faces limitations in scaling to multiple tasks due to computational costs. In this study, we propose a task addition approach for GATE to improve performance on target tasks with limited data while minimizing computational complexity. It is achieved through supervised multi-task pre-training on a large dataset, followed by the addition and training of task-specific modules for each target task. Our experiments demonstrate the superior performance of the task addition strategy for GATE over conventional multi-task methods, with comparable computational costs.
Multitask Extension of Geometrically Aligned Transfer Encoder
May 03, 2024



Abstract:Molecular datasets often suffer from a lack of data. It is well-known that gathering data is difficult due to the complexity of experimentation or simulation involved. Here, we leverage mutual information across different tasks in molecular data to address this issue. We extend an algorithm that utilizes the geometric characteristics of the encoding space, known as the Geometrically Aligned Transfer Encoder (GATE), to a multi-task setup. Thus, we connect multiple molecular tasks by aligning the curved coordinates onto locally flat coordinates, ensuring the flow of information from source tasks to support performance on target data.
Identification and validation of Triamcinolone and Gallopamil as treatments for early COVID-19 via an in silico repurposing pipeline
Jul 05, 2021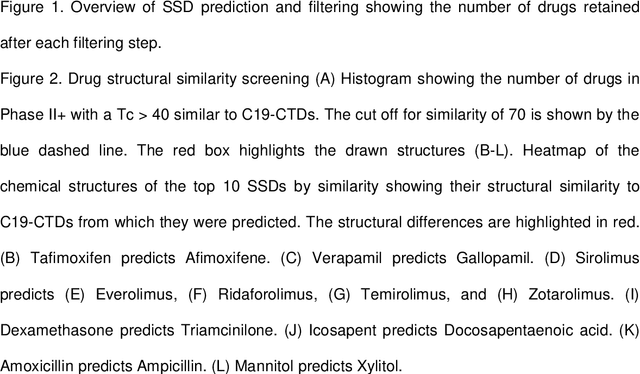
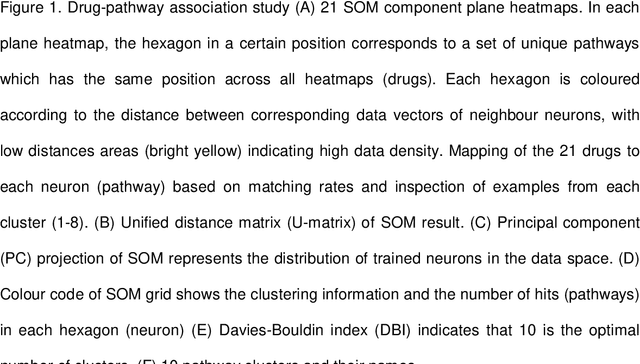
Abstract:SARS-CoV-2, the causative virus of COVID-19 continues to cause an ongoing global pandemic. Therapeutics are still needed to treat mild and severe COVID-19. Drug repurposing provides an opportunity to deploy drugs for COVID-19 more rapidly than developing novel therapeutics. Some existing drugs have shown promise for treating COVID-19 in clinical trials. This in silico study uses structural similarity to clinical trial drugs to identify two drugs with potential applications to treat early COVID-19. We apply in silico validation to suggest a possible mechanism of action for both. Triamcinolone is a corticosteroid structurally similar to Dexamethasone. Gallopamil is a calcium channel blocker structurally similar to Verapamil. We propose that both these drugs could be useful to treat early COVID-19 infection due to the proximity of their targets within a SARS-CoV-2-induced protein-protein interaction network to kinases active in early infection, and the APOA1 protein which is linked to the spread of COVID-19.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge