Michael Milham
NKI
A streamable large-scale clinical EEG dataset for Deep Learning
Apr 13, 2022


Abstract:Deep Learning has revolutionized various fields, including Computer Vision, Natural Language Processing, as well as Biomedical research. Within the field of neuroscience, specifically in electrophysiological neuroimaging, researchers are starting to explore leveraging deep learning to make predictions on their data without extensive feature engineering. The availability of large-scale datasets is a crucial aspect of allowing the experimentation of Deep Learning models. We are publishing the first large-scale clinical EEG dataset that simplifies data access and management for Deep Learning. This dataset contains eyes-closed EEG data prepared from a collection of 1,574 juvenile participants from the Healthy Brain Network. We demonstrate a use case integrating this framework, and discuss why providing such neuroinformatics infrastructure to the community is critical for future scientific discoveries.
Deep Convolutional Neural Network Applied to Electroencephalography: Raw Data vs Spectral Features
May 11, 2021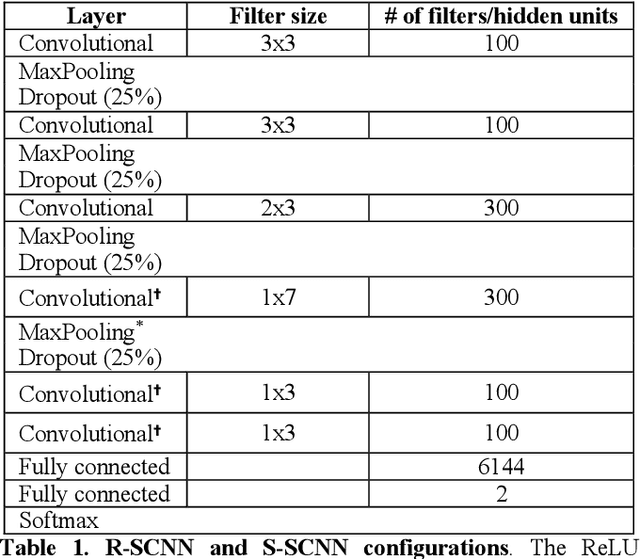

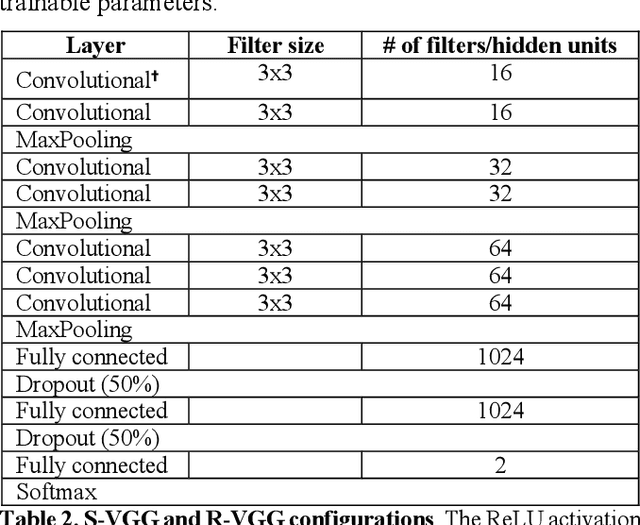
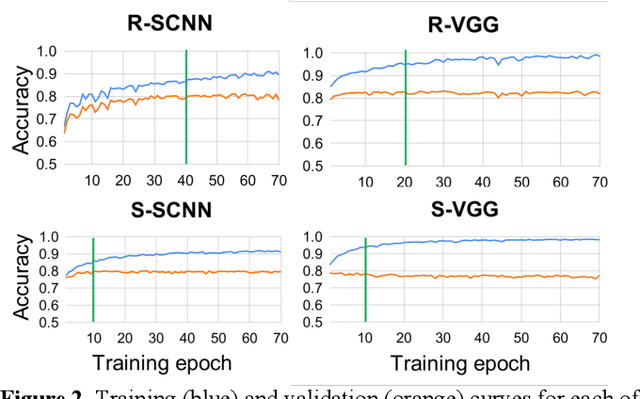
Abstract:The success of deep learning in computer vision has inspired the scientific community to explore new analysis methods. Within the field of neuroscience, specifically in electrophysiological neuroimaging, researchers are starting to explore leveraging deep learning to make predictions on their data without extensive feature engineering. This paper compares deep learning using minimally processed EEG raw data versus deep learning using EEG spectral features using two different deep convolutional neural architectures. One of them from Putten et al. (2018) is tailored to process raw data; the other was derived from the VGG16 vision network (Simonyan and Zisserman, 2015) which is designed to process EEG spectral features. We apply them to classify sex on 24-channel EEG from a large corpus of 1,574 participants. Not only do we improve on state-of-the-art classification performance for this type of classification problem, but we also show that in all cases, raw data classification leads to superior performance as compared to spectral EEG features. Interestingly we show that the neural network tailored to process EEG spectral features has increased performance when applied to raw data classification. Our approach suggests that the same convolutional networks used to process EEG spectral features yield superior performance when applied to EEG raw data.
Deriving reproducible biomarkers from multi-site resting-state data: An Autism-based example
Nov 18, 2016
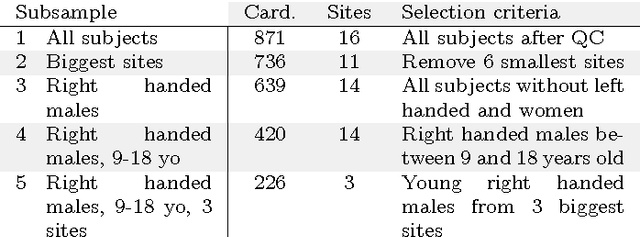
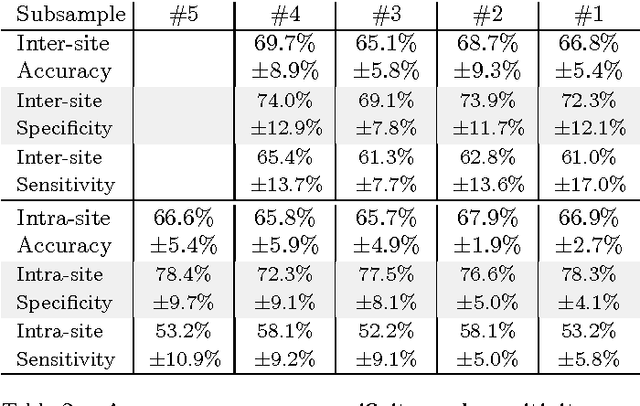

Abstract:Resting-state functional Magnetic Resonance Imaging (R-fMRI) holds the promise to reveal functional biomarkers of neuropsychiatric disorders. However, extracting such biomarkers is challenging for complex multi-faceted neuropatholo-gies, such as autism spectrum disorders. Large multi-site datasets increase sample sizes to compensate for this complexity, at the cost of uncontrolled heterogeneity. This heterogeneity raises new challenges, akin to those face in realistic diagnostic applications. Here, we demonstrate the feasibility of inter-site classification of neuropsychiatric status, with an application to the Autism Brain Imaging Data Exchange (ABIDE) database, a large (N=871) multi-site autism dataset. For this purpose, we investigate pipelines that extract the most predictive biomarkers from the data. These R-fMRI pipelines build participant-specific connectomes from functionally-defined brain areas. Connectomes are then compared across participants to learn patterns of connectivity that differentiate typical controls from individuals with autism. We predict this neuropsychiatric status for participants from the same acquisition sites or different, unseen, ones. Good choices of methods for the various steps of the pipeline lead to 67% prediction accuracy on the full ABIDE data, which is significantly better than previously reported results. We perform extensive validation on multiple subsets of the data defined by different inclusion criteria. These enables detailed analysis of the factors contributing to successful connectome-based prediction. First, prediction accuracy improves as we include more subjects, up to the maximum amount of subjects available. Second, the definition of functional brain areas is of paramount importance for biomarker discovery: brain areas extracted from large R-fMRI datasets outperform reference atlases in the classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge