Arnaud Delorme
EEG Foundation Challenge: From Cross-Task to Cross-Subject EEG Decoding
Jun 23, 2025Abstract:Current electroencephalogram (EEG) decoding models are typically trained on small numbers of subjects performing a single task. Here, we introduce a large-scale, code-submission-based competition comprising two challenges. First, the Transfer Challenge asks participants to build and test a model that can zero-shot decode new tasks and new subjects from their EEG data. Second, the Psychopathology factor prediction Challenge asks participants to infer subject measures of mental health from EEG data. For this, we use an unprecedented, multi-terabyte dataset of high-density EEG signals (128 channels) recorded from over 3,000 child to young adult subjects engaged in multiple active and passive tasks. We provide several tunable neural network baselines for each of these two challenges, including a simple network and demographic-based regression models. Developing models that generalise across tasks and individuals will pave the way for ML network architectures capable of adapting to EEG data collected from diverse tasks and individuals. Similarly, predicting mental health-relevant personality trait values from EEG might identify objective biomarkers useful for clinical diagnosis and design of personalised treatment for psychological conditions. Ultimately, the advances spurred by this challenge could contribute to the development of computational psychiatry and useful neurotechnology, and contribute to breakthroughs in both fundamental neuroscience and applied clinical research.
* Approved at Neurips Competition track. webpage: https://eeg2025.github.io/
Quantifying Data Requirements for EEG Independent Component Analysis Using AMICA
Jun 11, 2025Abstract:Independent Component Analysis (ICA) is an important step in EEG processing for a wide-ranging set of applications. However, ICA requires well-designed studies and data collection practices to yield optimal results. Past studies have focused on quantitative evaluation of the differences in quality produced by different ICA algorithms as well as different configurations of parameters for AMICA, a multimodal ICA algorithm that is considered the benchmark against which other algorithms are measured. Here, the effect of the data quantity versus the number of channels on decomposition quality is explored. AMICA decompositions were run on a 71 channel dataset with 13 subjects while randomly subsampling data to correspond to specific ratios of the number of frames in a dataset to the channel count. Decomposition quality was evaluated for the varying quantities of data using measures of mutual information reduction (MIR) and the near dipolarity of components. We also note that an asymptotic trend can be seen in the increase of MIR and a general increasing trend in near dipolarity with increasing data, but no definitive plateau in these metrics was observed, suggesting that the benefits of collecting additional EEG data may extend beyond common heuristic thresholds and continue to enhance decomposition quality.
Automatic EEG Independent Component Classification Using ICLabel in Python
Nov 20, 2024Abstract:ICLabel is an important plug-in function in EEGLAB, the most widely used software for EEG data processing. A powerful approach to automated processing of EEG data involves decomposing the data by Independent Component Analysis (ICA) and then classifying the resulting independent components (ICs) using ICLabel. While EEGLAB pipelines support high-performance computing (HPC) platforms running the open-source Octave interpreter, the ICLabel plug-in is incompatible with Octave because of its specialized neural network architecture. To enhance cross-platform compatibility, we developed a Python version of ICLabel that uses standard EEGLAB data structures. We compared ICLabel MATLAB and Python implementations to data from 14 subjects. ICLabel returns the likelihood of classification in 7 classes of components for each ICA component. The returned IC classifications were virtually identical between Python and MATLAB, with differences in classification percentage below 0.001%.
Deep learning applied to EEG data with different montages using spatial attention
Oct 16, 2023Abstract:The ability of Deep Learning to process and extract relevant information in complex brain dynamics from raw EEG data has been demonstrated in various recent works. Deep learning models, however, have also been shown to perform best on large corpora of data. When processing EEG, a natural approach is to combine EEG datasets from different experiments to train large deep-learning models. However, most EEG experiments use custom channel montages, requiring the data to be transformed into a common space. Previous methods have used the raw EEG signal to extract features of interest and focused on using a common feature space across EEG datasets. While this is a sensible approach, it underexploits the potential richness of EEG raw data. Here, we explore using spatial attention applied to EEG electrode coordinates to perform channel harmonization of raw EEG data, allowing us to train deep learning on EEG data using different montages. We test this model on a gender classification task. We first show that spatial attention increases model performance. Then, we show that a deep learning model trained on data using different channel montages performs significantly better than deep learning models trained on fixed 23- and 128-channel data montages.
An Exploration of Optimal Parameters for Efficient Blind Source Separation of EEG Recordings Using AMICA
Sep 27, 2023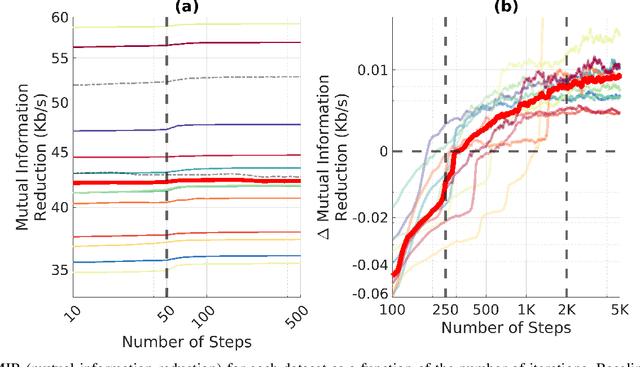
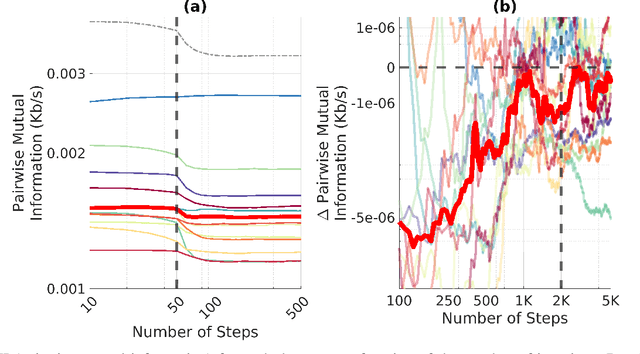
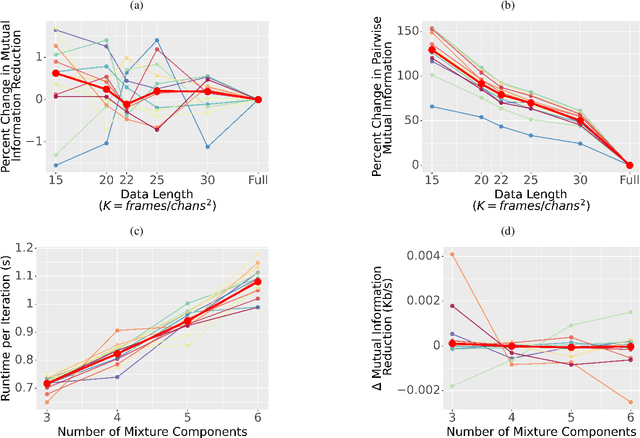
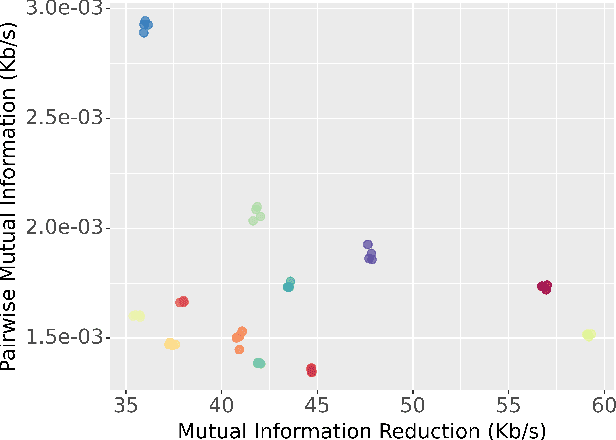
Abstract:EEG continues to find a multitude of uses in both neuroscience research and medical practice, and independent component analysis (ICA) continues to be an important tool for analyzing EEG. A multitude of ICA algorithms for EEG decomposition exist, and in the past, their relative effectiveness has been studied. AMICA is considered the benchmark against which to compare the performance of other ICA algorithms for EEG decomposition. AMICA exposes many parameters to the user to allow for precise control of the decomposition. However, several of the parameters currently tend to be set according to "rules of thumb" shared in the EEG community. Here, AMICA decompositions are run on data from a collection of subjects while varying certain key parameters. The running time and quality of decompositions are analyzed based on two metrics: Pairwise Mutual Information (PMI) and Mutual Information Reduction (MIR). Recommendations for selecting starting values for parameters are presented.
A Framework to Evaluate Independent Component Analysis applied to EEG signal: testing on the Picard algorithm
Oct 16, 2022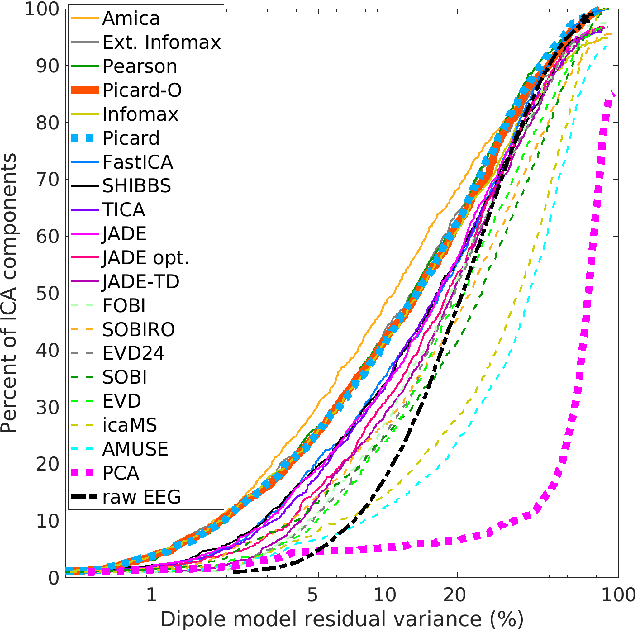
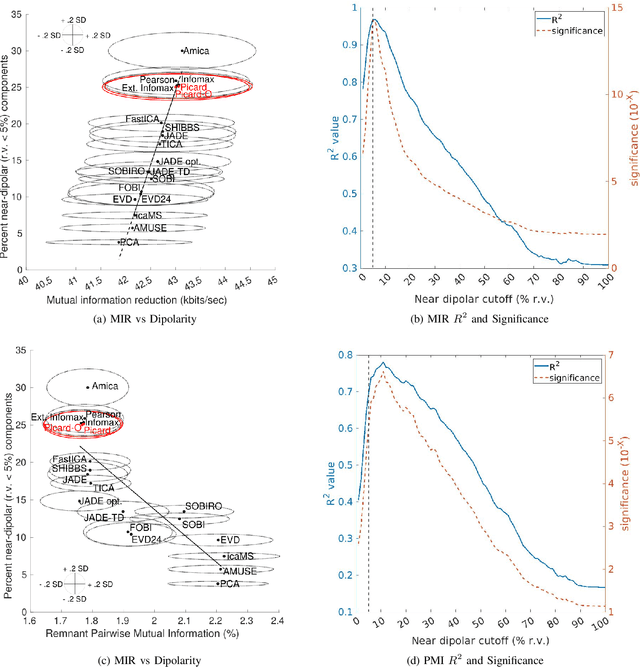
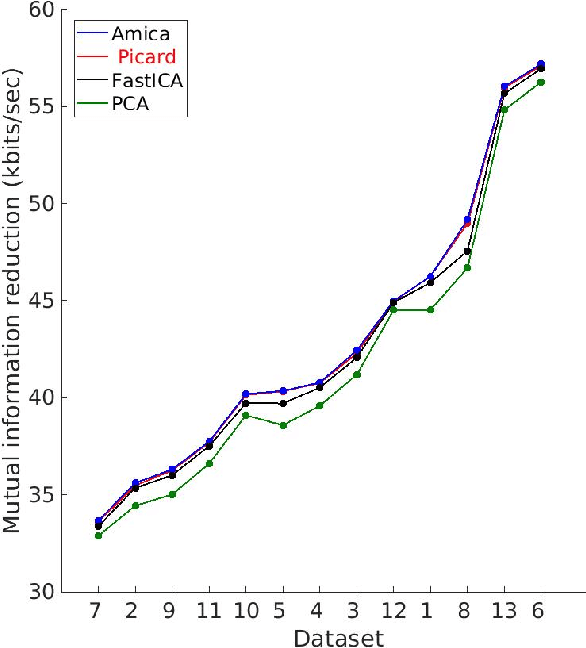
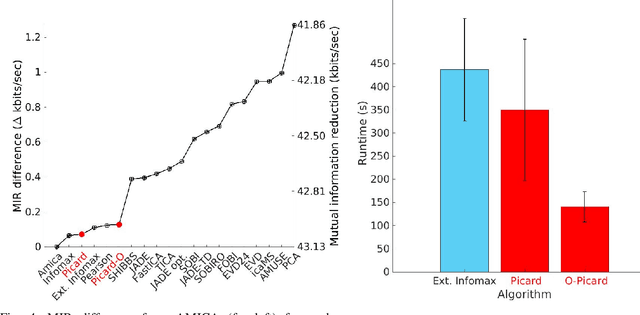
Abstract:Independent component analysis (ICA), is a blind source separation method that is becoming increasingly used to separate brain and non-brain related activities in electroencephalographic (EEG) and other electrophysiological recordings. It can be used to extract effective brain source activities and estimate their cortical source areas, and is commonly used in machine learning applications to classify EEG artifacts. Previously, we compared results of decomposing 13 71-channel scalp EEG datasets using 22 ICA and other blind source separation (BSS) algorithms. We are now making this framework available to the scientific community and, in the process of its release are testing a recent ICA algorithm (Picard) not included in the previous assay. Our test framework uses three main metrics to assess BSS performance: Pairwise Mutual Information (PMI) between scalp channel pairs; PMI remaining between component pairs after decomposition; and, the complete (not pairwise) Mutual Information Reduction (MIR) produced by each algorithm. We also measure the "dipolarity" of the scalp projection maps for the decomposed component, defined by the number of components whose scalp projection maps nearly match the projection of a single equivalent dipole. Within this framework, Picard performed similarly to Infomax ICA. This is not surprising since Picard is a type of Infomax algorithm that uses the L-BFGS method for faster convergence, in contrast to Infomax and Extended Infomax (runica) which use gradient descent. Our results show that Picard performs similarly to Infomax and, likewise, better than other BSS algorithms, excepting the more computationally complex AMICA. We have released the source code of our framework and the test data through GitHub.
A streamable large-scale clinical EEG dataset for Deep Learning
Apr 13, 2022


Abstract:Deep Learning has revolutionized various fields, including Computer Vision, Natural Language Processing, as well as Biomedical research. Within the field of neuroscience, specifically in electrophysiological neuroimaging, researchers are starting to explore leveraging deep learning to make predictions on their data without extensive feature engineering. The availability of large-scale datasets is a crucial aspect of allowing the experimentation of Deep Learning models. We are publishing the first large-scale clinical EEG dataset that simplifies data access and management for Deep Learning. This dataset contains eyes-closed EEG data prepared from a collection of 1,574 juvenile participants from the Healthy Brain Network. We demonstrate a use case integrating this framework, and discuss why providing such neuroinformatics infrastructure to the community is critical for future scientific discoveries.
Assessing learned features of Deep Learning applied to EEG
Nov 08, 2021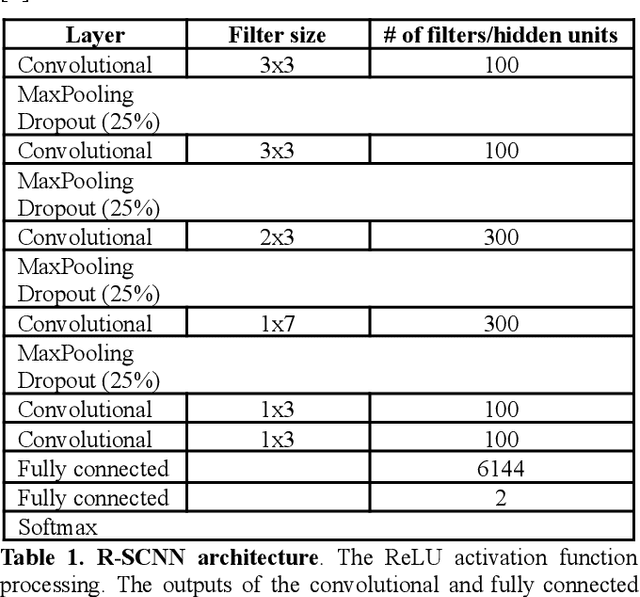
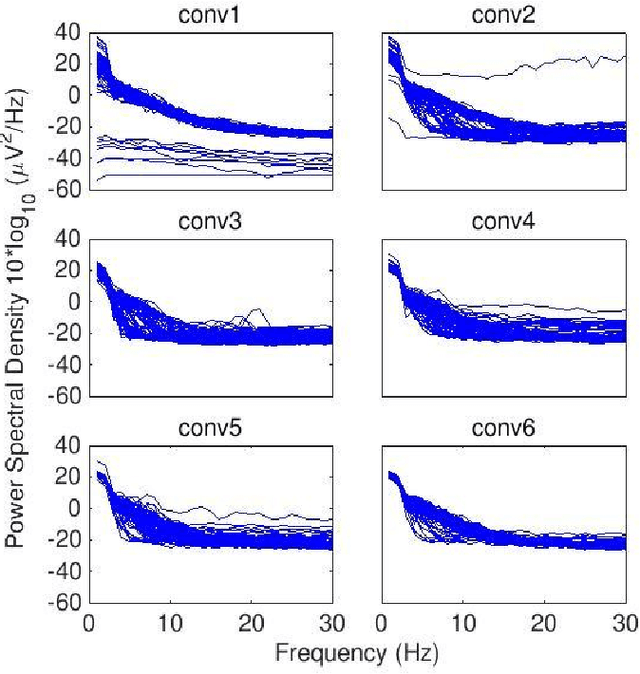
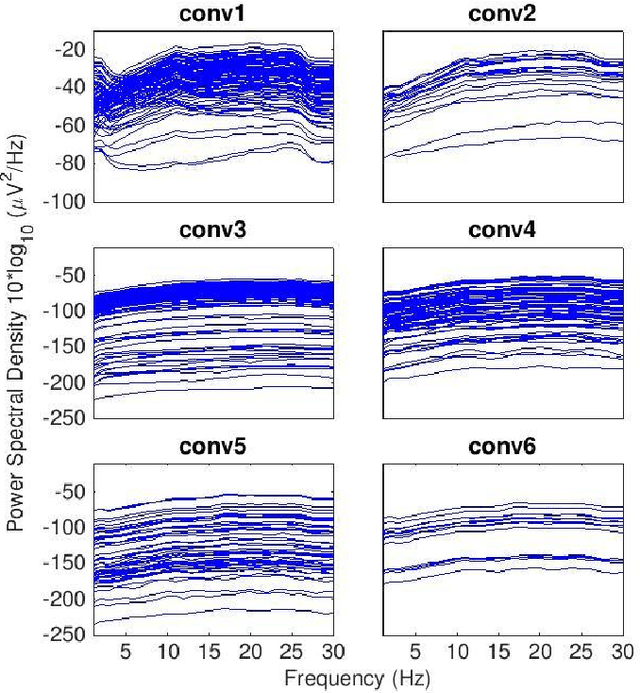
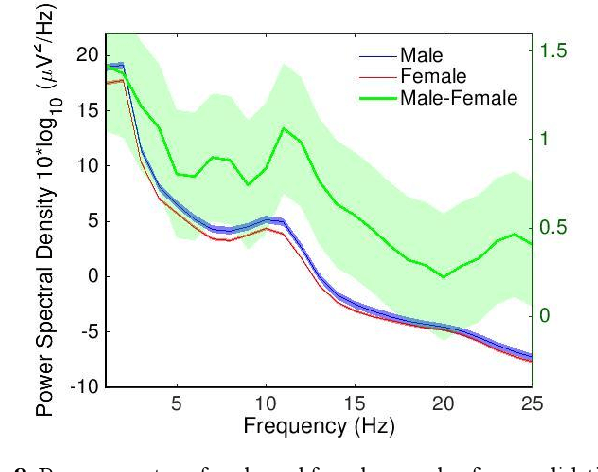
Abstract:Convolutional Neural Networks (CNNs) have achieved impressive performance on many computer vision related tasks, such as object detection, image recognition, image retrieval, etc. These achievements benefit from the CNNs' outstanding capability to learn discriminative features with deep layers of neuron structures and iterative training process. This has inspired the EEG research community to adopt CNN in performing EEG classification tasks. However, CNNs learned features are not immediately interpretable, causing a lack of understanding of the CNNs' internal working mechanism. To improve CNN interpretability, CNN visualization methods are applied to translate the internal features into visually perceptible patterns for qualitative analysis of CNN layers. Many CNN visualization methods have been proposed in the Computer Vision literature to interpret the CNN network structure, operation, and semantic concept, yet applications to EEG data analysis have been limited. In this work we use 3 different methods to extract EEG-relevant features from a CNN trained on raw EEG data: optimal samples for each classification category, activation maximization, and reverse convolution. We applied these methods to a high-performing Deep Learning model with state-of-the-art performance for an EEG sex classification task, and show that the model features a difference in the theta frequency band. We show that visualization of a CNN model can reveal interesting EEG results. Using these tools, EEG researchers using Deep Learning can better identify the learned EEG features, possibly identifying new class relevant biomarkers.
Deep Convolutional Neural Network Applied to Electroencephalography: Raw Data vs Spectral Features
May 11, 2021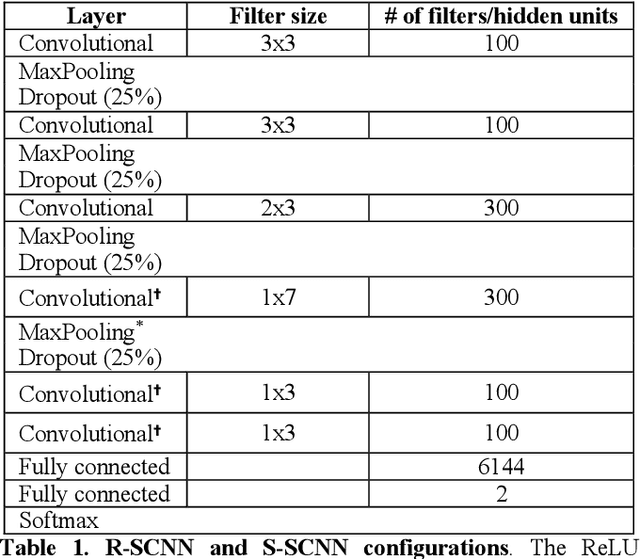

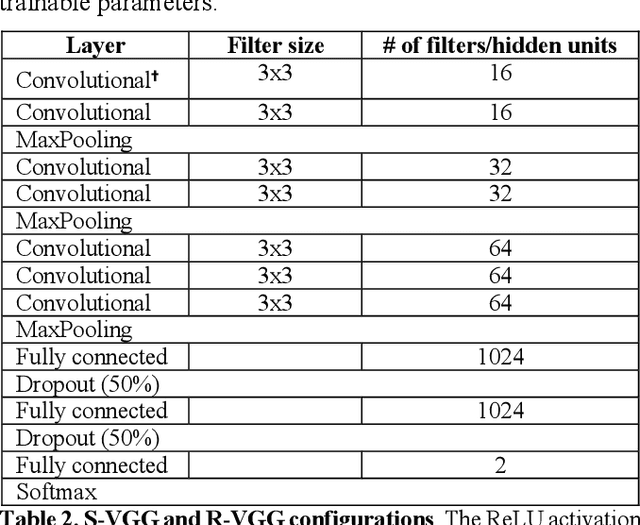
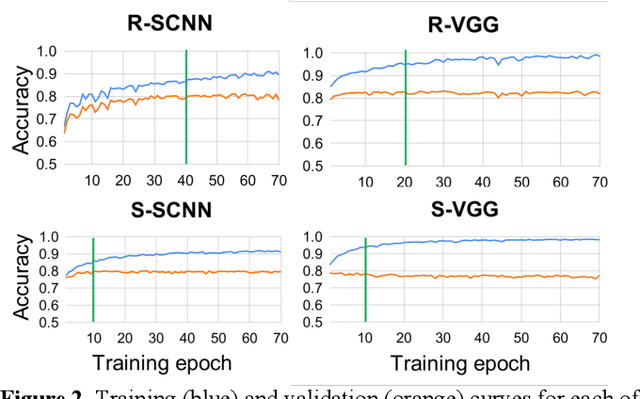
Abstract:The success of deep learning in computer vision has inspired the scientific community to explore new analysis methods. Within the field of neuroscience, specifically in electrophysiological neuroimaging, researchers are starting to explore leveraging deep learning to make predictions on their data without extensive feature engineering. This paper compares deep learning using minimally processed EEG raw data versus deep learning using EEG spectral features using two different deep convolutional neural architectures. One of them from Putten et al. (2018) is tailored to process raw data; the other was derived from the VGG16 vision network (Simonyan and Zisserman, 2015) which is designed to process EEG spectral features. We apply them to classify sex on 24-channel EEG from a large corpus of 1,574 participants. Not only do we improve on state-of-the-art classification performance for this type of classification problem, but we also show that in all cases, raw data classification leads to superior performance as compared to spectral EEG features. Interestingly we show that the neural network tailored to process EEG spectral features has increased performance when applied to raw data classification. Our approach suggests that the same convolutional networks used to process EEG spectral features yield superior performance when applied to EEG raw data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge