Gwenevere Frank
Quantifying Data Requirements for EEG Independent Component Analysis Using AMICA
Jun 11, 2025Abstract:Independent Component Analysis (ICA) is an important step in EEG processing for a wide-ranging set of applications. However, ICA requires well-designed studies and data collection practices to yield optimal results. Past studies have focused on quantitative evaluation of the differences in quality produced by different ICA algorithms as well as different configurations of parameters for AMICA, a multimodal ICA algorithm that is considered the benchmark against which other algorithms are measured. Here, the effect of the data quantity versus the number of channels on decomposition quality is explored. AMICA decompositions were run on a 71 channel dataset with 13 subjects while randomly subsampling data to correspond to specific ratios of the number of frames in a dataset to the channel count. Decomposition quality was evaluated for the varying quantities of data using measures of mutual information reduction (MIR) and the near dipolarity of components. We also note that an asymptotic trend can be seen in the increase of MIR and a general increasing trend in near dipolarity with increasing data, but no definitive plateau in these metrics was observed, suggesting that the benefits of collecting additional EEG data may extend beyond common heuristic thresholds and continue to enhance decomposition quality.
An Exploration of Optimal Parameters for Efficient Blind Source Separation of EEG Recordings Using AMICA
Sep 27, 2023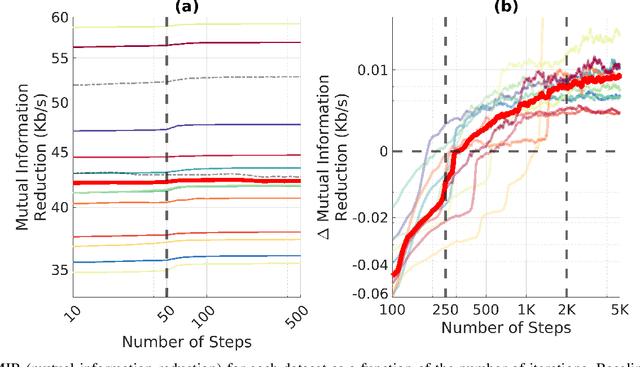
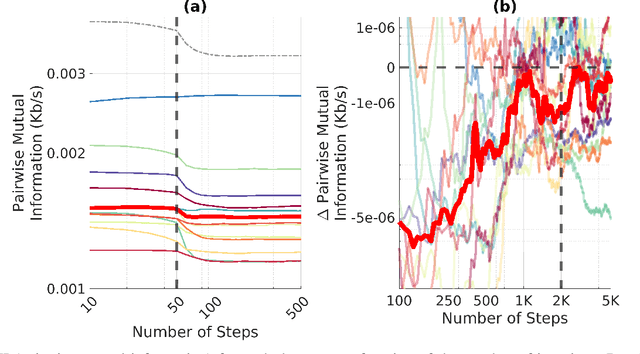
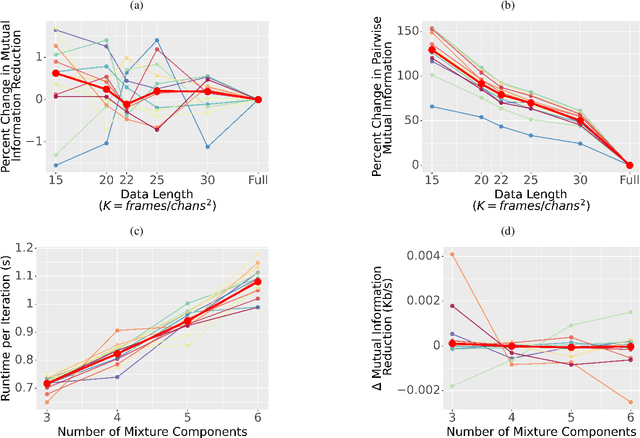
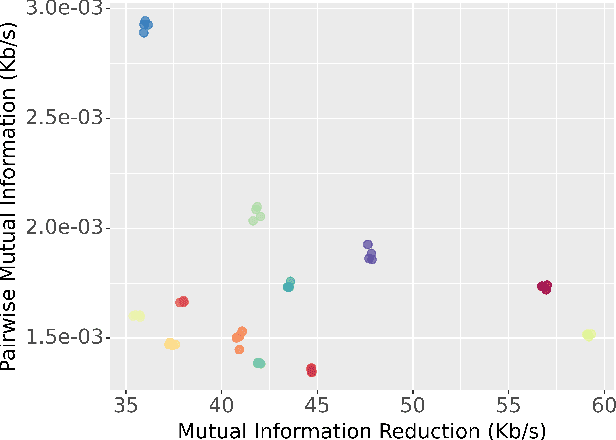
Abstract:EEG continues to find a multitude of uses in both neuroscience research and medical practice, and independent component analysis (ICA) continues to be an important tool for analyzing EEG. A multitude of ICA algorithms for EEG decomposition exist, and in the past, their relative effectiveness has been studied. AMICA is considered the benchmark against which to compare the performance of other ICA algorithms for EEG decomposition. AMICA exposes many parameters to the user to allow for precise control of the decomposition. However, several of the parameters currently tend to be set according to "rules of thumb" shared in the EEG community. Here, AMICA decompositions are run on data from a collection of subjects while varying certain key parameters. The running time and quality of decompositions are analyzed based on two metrics: Pairwise Mutual Information (PMI) and Mutual Information Reduction (MIR). Recommendations for selecting starting values for parameters are presented.
A Framework to Evaluate Independent Component Analysis applied to EEG signal: testing on the Picard algorithm
Oct 16, 2022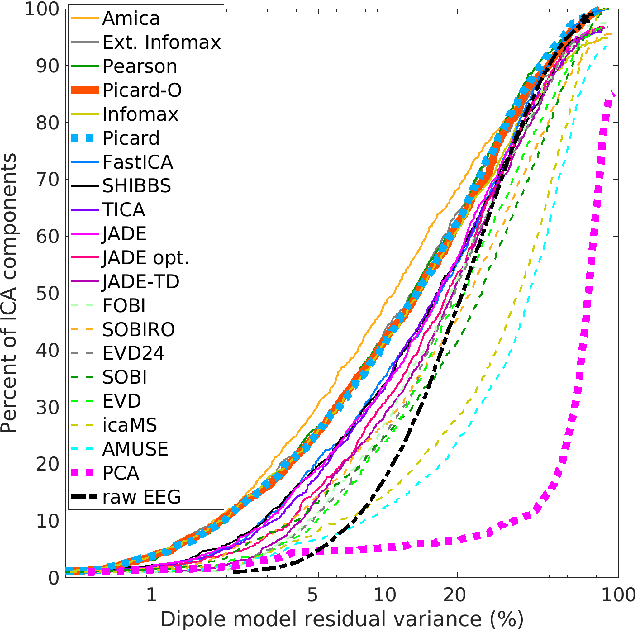
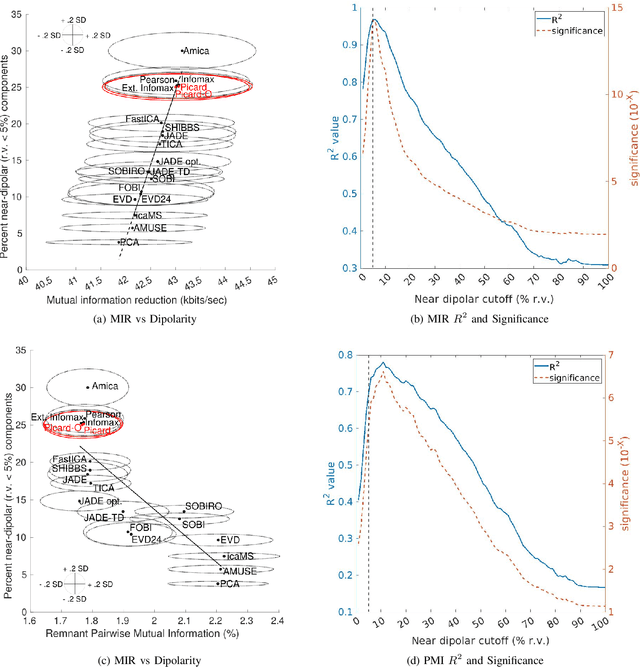
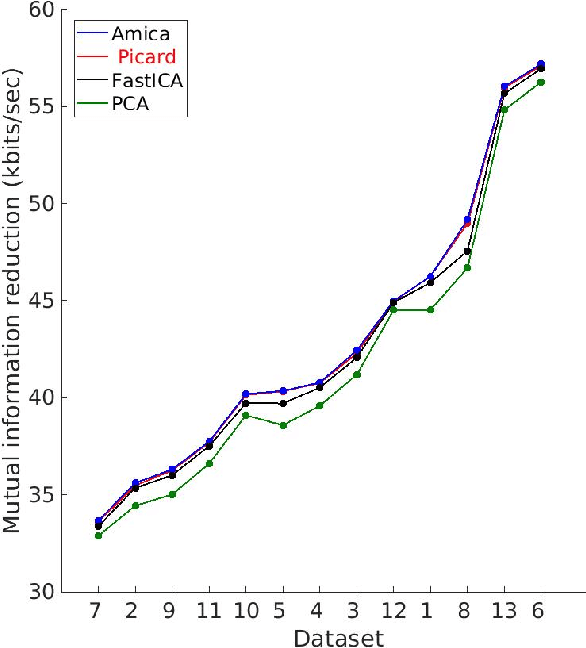
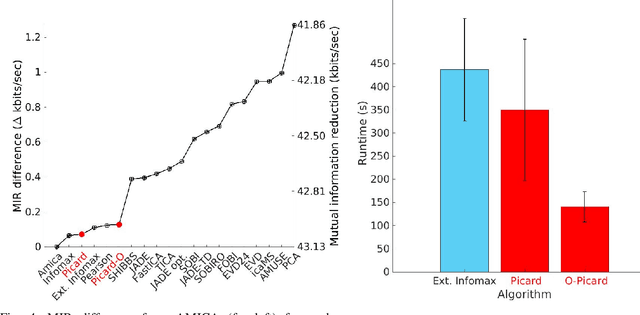
Abstract:Independent component analysis (ICA), is a blind source separation method that is becoming increasingly used to separate brain and non-brain related activities in electroencephalographic (EEG) and other electrophysiological recordings. It can be used to extract effective brain source activities and estimate their cortical source areas, and is commonly used in machine learning applications to classify EEG artifacts. Previously, we compared results of decomposing 13 71-channel scalp EEG datasets using 22 ICA and other blind source separation (BSS) algorithms. We are now making this framework available to the scientific community and, in the process of its release are testing a recent ICA algorithm (Picard) not included in the previous assay. Our test framework uses three main metrics to assess BSS performance: Pairwise Mutual Information (PMI) between scalp channel pairs; PMI remaining between component pairs after decomposition; and, the complete (not pairwise) Mutual Information Reduction (MIR) produced by each algorithm. We also measure the "dipolarity" of the scalp projection maps for the decomposed component, defined by the number of components whose scalp projection maps nearly match the projection of a single equivalent dipole. Within this framework, Picard performed similarly to Infomax ICA. This is not surprising since Picard is a type of Infomax algorithm that uses the L-BFGS method for faster convergence, in contrast to Infomax and Extended Infomax (runica) which use gradient descent. Our results show that Picard performs similarly to Infomax and, likewise, better than other BSS algorithms, excepting the more computationally complex AMICA. We have released the source code of our framework and the test data through GitHub.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge