Aiping Liu
Dual Graph Attention based Disentanglement Multiple Instance Learning for Brain Age Estimation
Mar 02, 2024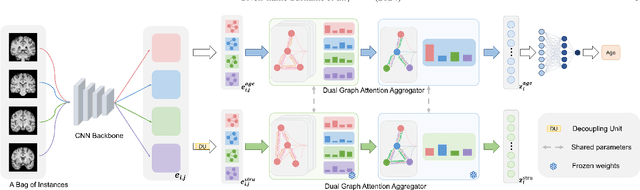
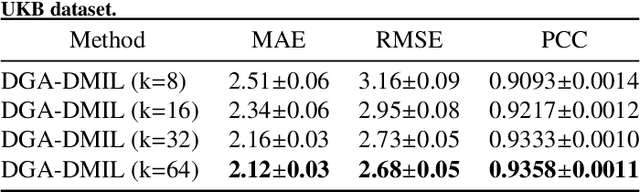
Abstract:Deep learning techniques have demonstrated great potential for accurately estimating brain age by analyzing Magnetic Resonance Imaging (MRI) data from healthy individuals. However, current methods for brain age estimation often directly utilize whole input images, overlooking two important considerations: 1) the heterogeneous nature of brain aging, where different brain regions may degenerate at different rates, and 2) the existence of age-independent redundancies in brain structure. To overcome these limitations, we propose a Dual Graph Attention based Disentanglement Multi-instance Learning (DGA-DMIL) framework for improving brain age estimation. Specifically, the 3D MRI data, treated as a bag of instances, is fed into a 2D convolutional neural network backbone, to capture the unique aging patterns in MRI. A dual graph attention aggregator is then proposed to learn the backbone features by exploiting the intra- and inter-instance relationships. Furthermore, a disentanglement branch is introduced to separate age-related features from age-independent structural representations to ameliorate the interference of redundant information on age prediction. To verify the effectiveness of the proposed framework, we evaluate it on two datasets, UK Biobank and ADNI, containing a total of 35,388 healthy individuals. Our proposed model demonstrates exceptional accuracy in estimating brain age, achieving a remarkable mean absolute error of 2.12 years in the UK Biobank. The results establish our approach as state-of-the-art compared to other competing brain age estimation models. In addition, the instance contribution scores identify the varied importance of brain areas for aging prediction, which provides deeper insights into the understanding of brain aging.
Data Augmentation for Seizure Prediction with Generative Diffusion Model
Jun 14, 2023Abstract:Objective: Seizure prediction is of great importance to improve the life of patients. The focal point is to distinguish preictal states from interictal ones. With the development of machine learning, seizure prediction methods have achieved significant progress. However, the severe imbalance problem between preictal and interictal data still poses a great challenge, restricting the performance of classifiers. Data augmentation is an intuitive way to solve this problem. Existing data augmentation methods generate samples by overlapping or recombining data. The distribution of generated samples is limited by original data, because such transformations cannot fully explore the feature space and offer new information. As the epileptic EEG representation varies among seizures, these generated samples cannot provide enough diversity to achieve high performance on a new seizure. As a consequence, we propose a novel data augmentation method with diffusion model called DiffEEG. Methods: Diffusion models are a class of generative models that consist of two processes. Specifically, in the diffusion process, the model adds noise to the input EEG sample step by step and converts the noisy sample into output random noise, exploring the distribution of data by minimizing the loss between the output and the noise added. In the denoised process, the model samples the synthetic data by removing the noise gradually, diffusing the data distribution to outward areas and narrowing the distance between different clusters. Results: We compared DiffEEG with existing methods, and integrated them into three representative classifiers. The experiments indicate that DiffEEG could further improve the performance and shows superiority to existing methods. Conclusion: This paper proposes a novel and effective method to solve the imbalanced problem and demonstrates the effectiveness and generality of our method.
PanFlowNet: A Flow-Based Deep Network for Pan-sharpening
May 16, 2023Abstract:Pan-sharpening aims to generate a high-resolution multispectral (HRMS) image by integrating the spectral information of a low-resolution multispectral (LRMS) image with the texture details of a high-resolution panchromatic (PAN) image. It essentially inherits the ill-posed nature of the super-resolution (SR) task that diverse HRMS images can degrade into an LRMS image. However, existing deep learning-based methods recover only one HRMS image from the LRMS image and PAN image using a deterministic mapping, thus ignoring the diversity of the HRMS image. In this paper, to alleviate this ill-posed issue, we propose a flow-based pan-sharpening network (PanFlowNet) to directly learn the conditional distribution of HRMS image given LRMS image and PAN image instead of learning a deterministic mapping. Specifically, we first transform this unknown conditional distribution into a given Gaussian distribution by an invertible network, and the conditional distribution can thus be explicitly defined. Then, we design an invertible Conditional Affine Coupling Block (CACB) and further build the architecture of PanFlowNet by stacking a series of CACBs. Finally, the PanFlowNet is trained by maximizing the log-likelihood of the conditional distribution given a training set and can then be used to predict diverse HRMS images. The experimental results verify that the proposed PanFlowNet can generate various HRMS images given an LRMS image and a PAN image. Additionally, the experimental results on different kinds of satellite datasets also demonstrate the superiority of our PanFlowNet compared with other state-of-the-art methods both visually and quantitatively.
Model-Guided Multi-Contrast Deep Unfolding Network for MRI Super-resolution Reconstruction
Sep 15, 2022



Abstract:Magnetic resonance imaging (MRI) with high resolution (HR) provides more detailed information for accurate diagnosis and quantitative image analysis. Despite the significant advances, most existing super-resolution (SR) reconstruction network for medical images has two flaws: 1) All of them are designed in a black-box principle, thus lacking sufficient interpretability and further limiting their practical applications. Interpretable neural network models are of significant interest since they enhance the trustworthiness required in clinical practice when dealing with medical images. 2) most existing SR reconstruction approaches only use a single contrast or use a simple multi-contrast fusion mechanism, neglecting the complex relationships between different contrasts that are critical for SR improvement. To deal with these issues, in this paper, a novel Model-Guided interpretable Deep Unfolding Network (MGDUN) for medical image SR reconstruction is proposed. The Model-Guided image SR reconstruction approach solves manually designed objective functions to reconstruct HR MRI. We show how to unfold an iterative MGDUN algorithm into a novel model-guided deep unfolding network by taking the MRI observation matrix and explicit multi-contrast relationship matrix into account during the end-to-end optimization. Extensive experiments on the multi-contrast IXI dataset and BraTs 2019 dataset demonstrate the superiority of our proposed model.
Toward Open-World Electroencephalogram Decoding Via Deep Learning: A Comprehensive Survey
Dec 17, 2021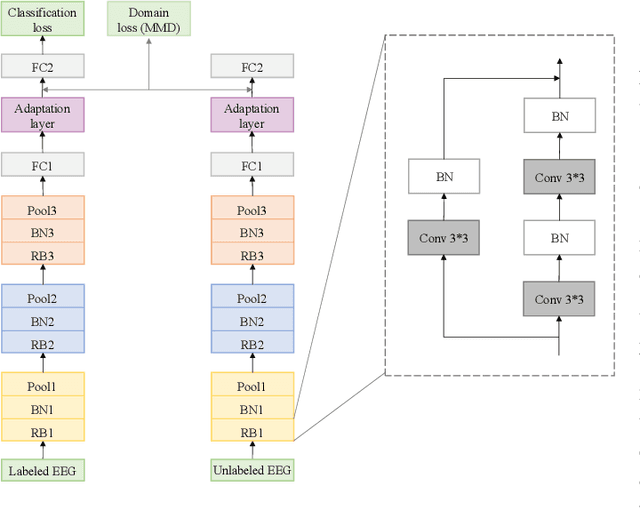
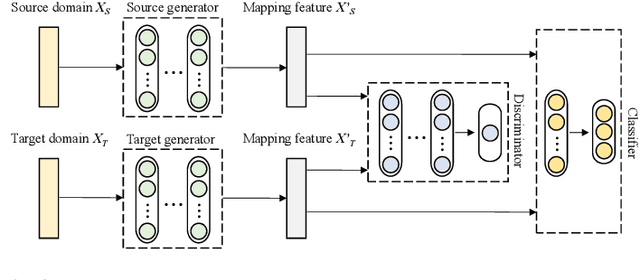
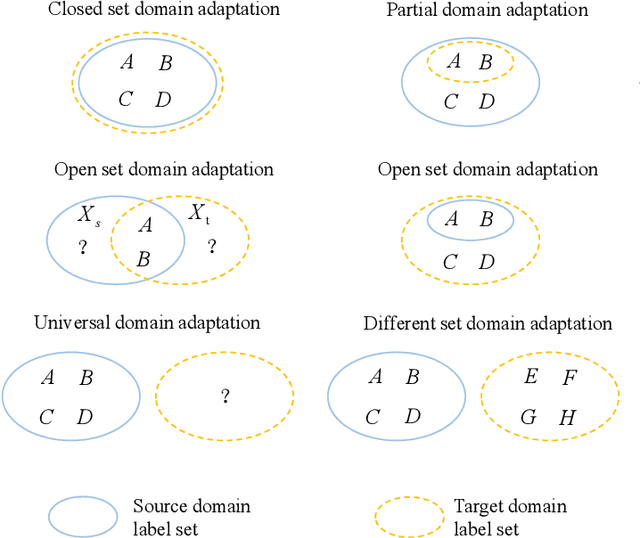
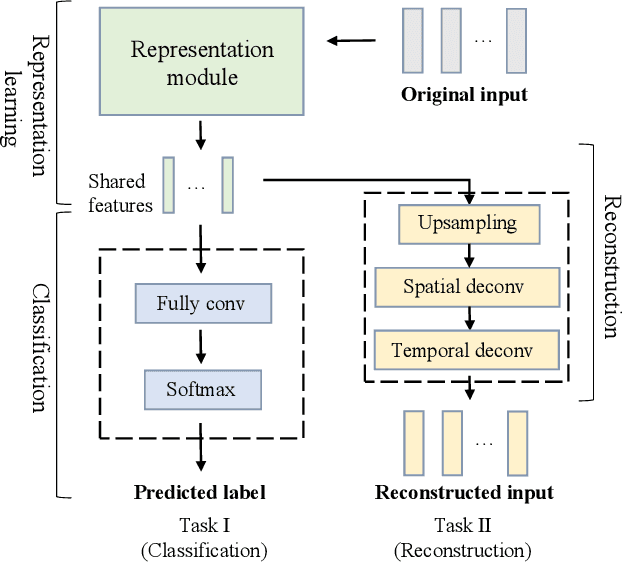
Abstract:Electroencephalogram (EEG) decoding aims to identify the perceptual, semantic, and cognitive content of neural processing based on non-invasively measured brain activity. Traditional EEG decoding methods have achieved moderate success when applied to data acquired in static, well-controlled lab environments. However, an open-world environment is a more realistic setting, where situations affecting EEG recordings can emerge unexpectedly, significantly weakening the robustness of existing methods. In recent years, deep learning (DL) has emerged as a potential solution for such problems due to its superior capacity in feature extraction. It overcomes the limitations of defining `handcrafted' features or features extracted using shallow architectures, but typically requires large amounts of costly, expertly-labelled data - something not always obtainable. Combining DL with domain-specific knowledge may allow for development of robust approaches to decode brain activity even with small-sample data. Although various DL methods have been proposed to tackle some of the challenges in EEG decoding, a systematic tutorial overview, particularly for open-world applications, is currently lacking. This article therefore provides a comprehensive survey of DL methods for open-world EEG decoding, and identifies promising research directions to inspire future studies for EEG decoding in real-world applications.
Automated assessment of disease severity of COVID-19 using artificial intelligence with synthetic chest CT
Dec 11, 2021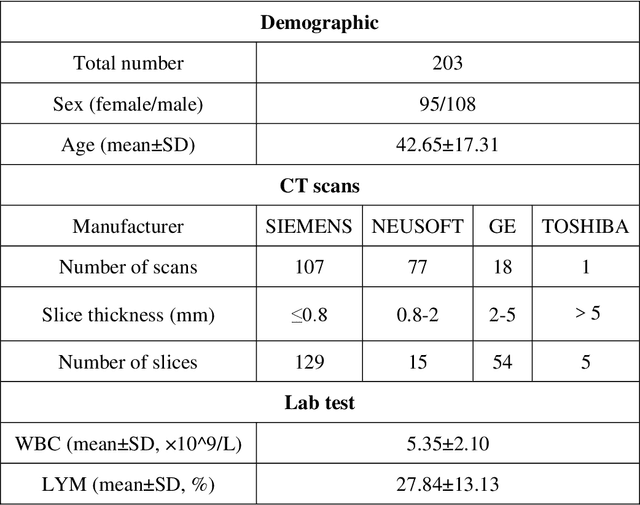
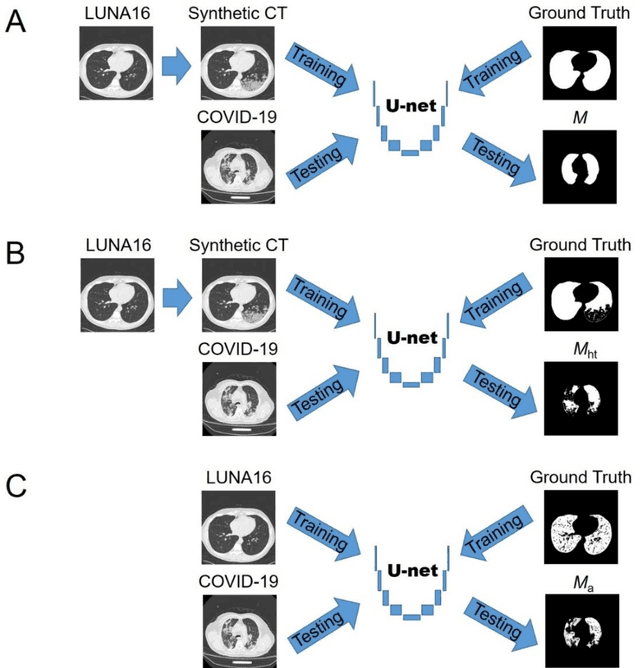
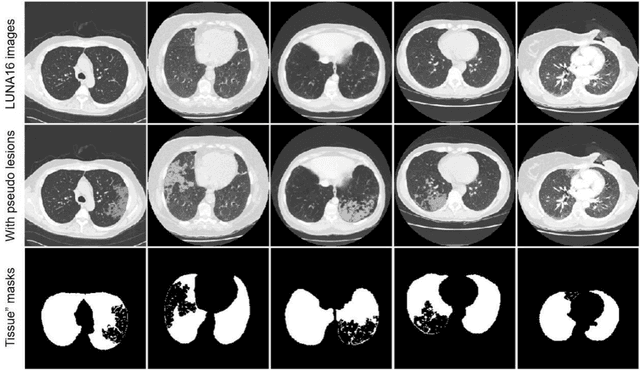
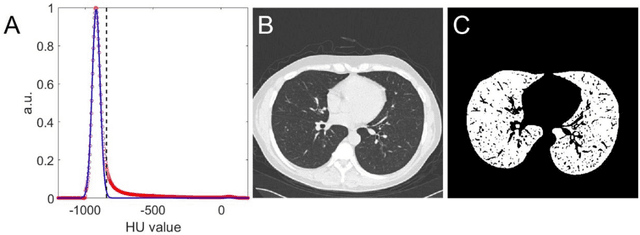
Abstract:Background: Triage of patients is important to control the pandemic of coronavirus disease 2019 (COVID-19), especially during the peak of the pandemic when clinical resources become extremely limited. Purpose: To develop a method that automatically segments and quantifies lung and pneumonia lesions with synthetic chest CT and assess disease severity in COVID-19 patients. Materials and Methods: In this study, we incorporated data augmentation to generate synthetic chest CT images using public available datasets (285 datasets from "Lung Nodule Analysis 2016"). The synthetic images and masks were used to train a 2D U-net neural network and tested on 203 COVID-19 datasets to generate lung and lesion segmentations. Disease severity scores (DL: damage load; DS: damage score) were calculated based on the segmentations. Correlations between DL/DS and clinical lab tests were evaluated using Pearson's method. A p-value < 0.05 was considered as statistical significant. Results: Automatic lung and lesion segmentations were compared with manual annotations. For lung segmentation, the median values of dice similarity coefficient, Jaccard index and average surface distance, were 98.56%, 97.15% and 0.49 mm, respectively. The same metrics for lesion segmentation were 76.95%, 62.54% and 2.36 mm, respectively. Significant (p << 0.05) correlations were found between DL/DS and percentage lymphocytes tests, with r-values of -0.561 and -0.501, respectively. Conclusion: An AI system that based on thoracic radiographic and data augmentation was proposed to segment lung and lesions in COVID-19 patients. Correlations between imaging findings and clinical lab tests suggested the value of this system as a potential tool to assess disease severity of COVID-19.
Unfolding Taylor's Approximations for Image Restoration
Sep 08, 2021



Abstract:Deep learning provides a new avenue for image restoration, which demands a delicate balance between fine-grained details and high-level contextualized information during recovering the latent clear image. In practice, however, existing methods empirically construct encapsulated end-to-end mapping networks without deepening into the rationality, and neglect the intrinsic prior knowledge of restoration task. To solve the above problems, inspired by Taylor's Approximations, we unfold Taylor's Formula to construct a novel framework for image restoration. We find the main part and the derivative part of Taylor's Approximations take the same effect as the two competing goals of high-level contextualized information and spatial details of image restoration respectively. Specifically, our framework consists of two steps, correspondingly responsible for the mapping and derivative functions. The former first learns the high-level contextualized information and the later combines it with the degraded input to progressively recover local high-order spatial details. Our proposed framework is orthogonal to existing methods and thus can be easily integrated with them for further improvement, and extensive experiments demonstrate the effectiveness and scalability of our proposed framework.
MLBF-Net: A Multi-Lead-Branch Fusion Network for Multi-Class Arrhythmia Classification Using 12-Lead ECG
Aug 17, 2020



Abstract:Automatic arrhythmia detection using 12-lead electrocardiogram (ECG) signal plays a critical role in early prevention and diagnosis of cardiovascular diseases. In the previous studies on automatic arrhythmia detection, most methods concatenated 12 leads of ECG into a matrix, and then input the matrix to a variety of feature extractors or deep neural networks for extracting useful information. Under such frameworks, these methods had the ability to extract comprehensive features (known as integrity) of 12-lead ECG since the information of each lead interacts with each other during training. However, the diverse lead-specific features (known as diversity) among 12 leads were neglected, causing inadequate information learning for 12-lead ECG. To maximize the information learning of multi-lead ECG, the information fusion of comprehensive features with integrity and lead-specific features with diversity should be taken into account. In this paper, we propose a novel Multi-Lead-Branch Fusion Network (MLBF-Net) architecture for arrhythmia classification by integrating multi-loss optimization to jointly learning diversity and integrity of multi-lead ECG. MLBF-Net is composed of three components: 1) multiple lead-specific branches for learning the diversity of multi-lead ECG; 2) cross-lead features fusion by concatenating the output feature maps of all branches for learning the integrity of multi-lead ECG; 3) multi-loss co-optimization for all the individual branches and the concatenated network. We demonstrate our MLBF-Net on China Physiological Signal Challenge 2018 which is an open 12-lead ECG dataset. The experimental results show that MLBF-Net obtains an average $F_1$ score of 0.855, reaching the highest arrhythmia classification performance. The proposed method provides a promising solution for multi-lead ECG analysis from an information fusion perspective.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge