Yidong Yang
Free-form Lesion Synthesis Using a Partial Convolution Generative Adversarial Network for Enhanced Deep Learning Liver Tumor Segmentation
Jun 18, 2022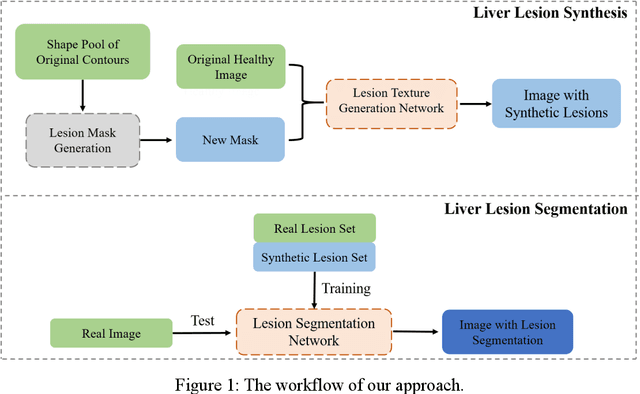
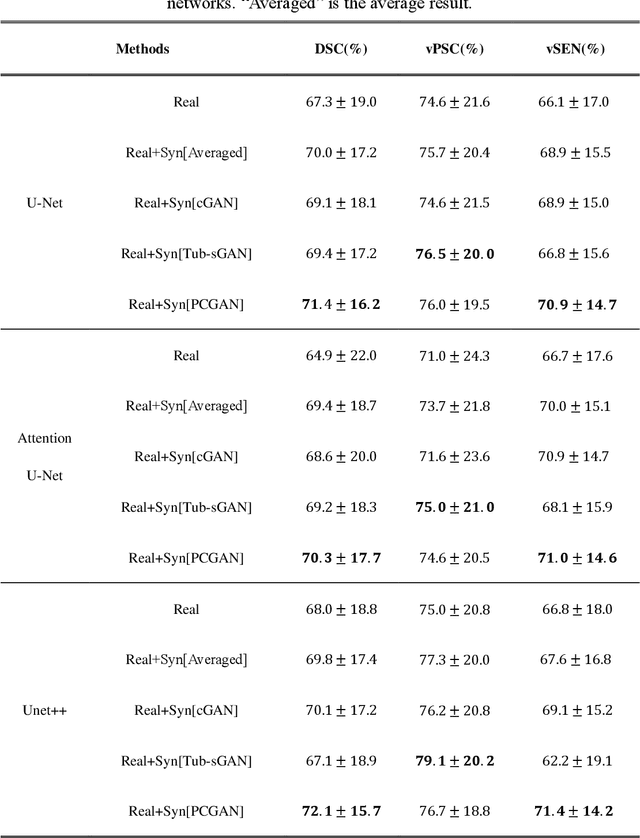

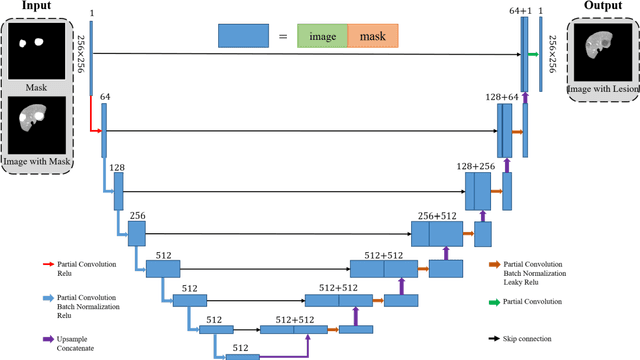
Abstract:Automatic deep learning segmentation models has been shown to improve both the segmentation efficiency and the accuracy. However, training a robust segmentation model requires considerably large labeled training samples, which may be impractical. This study aimed to develop a deep learning framework for generating synthetic lesions that can be used to enhance network training. The lesion synthesis network is a modified generative adversarial network (GAN). Specifically, we innovated a partial convolution strategy to construct an Unet-like generator. The discriminator is designed using Wasserstein GAN with gradient penalty and spectral normalization. A mask generation method based on principal component analysis was developed to model various lesion shapes. The generated masks are then converted into liver lesions through a lesion synthesis network. The lesion synthesis framework was evaluated for lesion textures, and the synthetic lesions were used to train a lesion segmentation network to further validate the effectiveness of this framework. All the networks are trained and tested on the public dataset from LITS. The synthetic lesions generated by the proposed approach have very similar histogram distributions compared to the real lesions for the two employed texture parameters, GLCM-energy and GLCM-correlation. The Kullback-Leibler divergence of GLCM-energy and GLCM-correlation were 0.01 and 0.10, respectively. Including the synthetic lesions in the tumor segmentation network improved the segmentation dice performance of U-Net significantly from 67.3% to 71.4% (p<0.05). Meanwhile, the volume precision and sensitivity improve from 74.6% to 76.0% (p=0.23) and 66.1% to 70.9% (p<0.01), respectively. The synthetic data significantly improves the segmentation performance.
Automated assessment of disease severity of COVID-19 using artificial intelligence with synthetic chest CT
Dec 11, 2021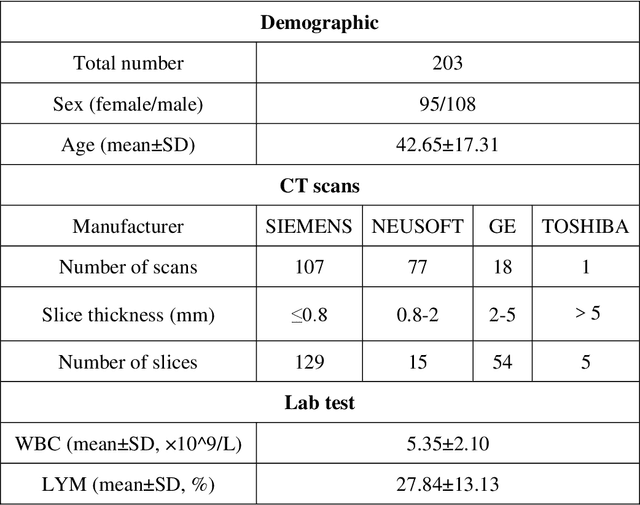
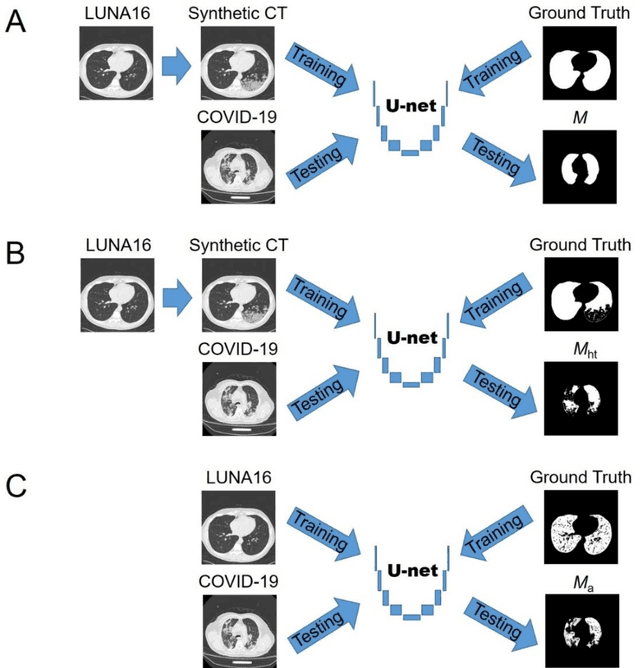
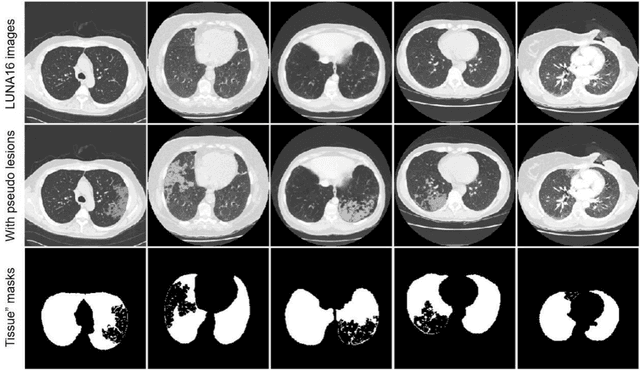
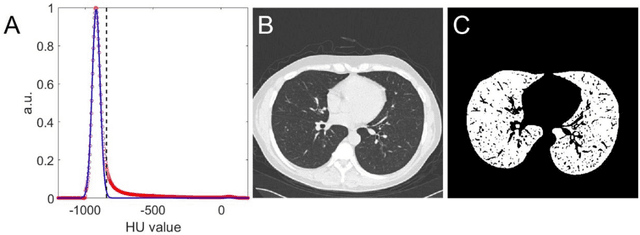
Abstract:Background: Triage of patients is important to control the pandemic of coronavirus disease 2019 (COVID-19), especially during the peak of the pandemic when clinical resources become extremely limited. Purpose: To develop a method that automatically segments and quantifies lung and pneumonia lesions with synthetic chest CT and assess disease severity in COVID-19 patients. Materials and Methods: In this study, we incorporated data augmentation to generate synthetic chest CT images using public available datasets (285 datasets from "Lung Nodule Analysis 2016"). The synthetic images and masks were used to train a 2D U-net neural network and tested on 203 COVID-19 datasets to generate lung and lesion segmentations. Disease severity scores (DL: damage load; DS: damage score) were calculated based on the segmentations. Correlations between DL/DS and clinical lab tests were evaluated using Pearson's method. A p-value < 0.05 was considered as statistical significant. Results: Automatic lung and lesion segmentations were compared with manual annotations. For lung segmentation, the median values of dice similarity coefficient, Jaccard index and average surface distance, were 98.56%, 97.15% and 0.49 mm, respectively. The same metrics for lesion segmentation were 76.95%, 62.54% and 2.36 mm, respectively. Significant (p << 0.05) correlations were found between DL/DS and percentage lymphocytes tests, with r-values of -0.561 and -0.501, respectively. Conclusion: An AI system that based on thoracic radiographic and data augmentation was proposed to segment lung and lesions in COVID-19 patients. Correlations between imaging findings and clinical lab tests suggested the value of this system as a potential tool to assess disease severity of COVID-19.
Unsupervised COVID-19 Lesion Segmentation in CT Using Cycle Consistent Generative Adversarial Network
Nov 23, 2021

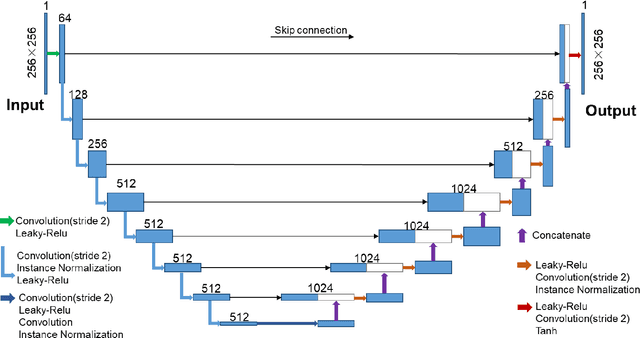
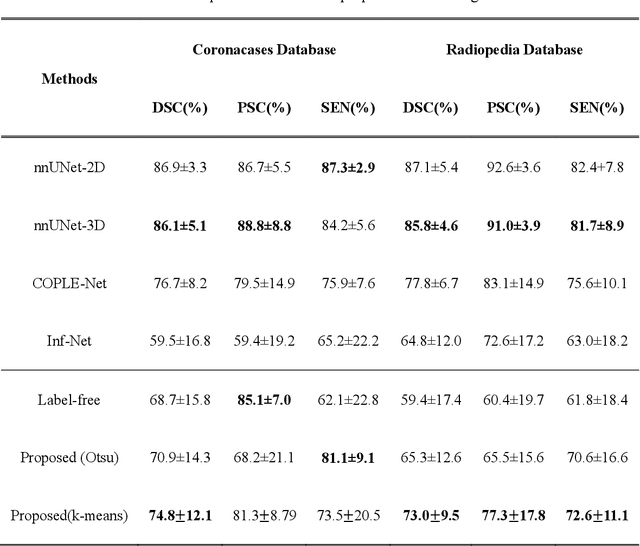
Abstract:COVID-19 has become a global pandemic and is still posing a severe health risk to the public. Accurate and efficient segmentation of pneumonia lesions in CT scans is vital for treatment decision-making. We proposed a novel unsupervised approach using cycle consistent generative adversarial network (cycle-GAN) which automates and accelerates the process of lesion delineation. The workflow includes lung volume segmentation, "synthetic" healthy lung generation, infected and healthy image subtraction, and binary lesion mask creation. The lung volume volume was firstly delineated using a pre-trained U-net and worked as the input for the later network. The cycle-GAN was developed to generate synthetic "healthy" lung CT images from infected lung images. After that, the pneumonia lesions are extracted by subtracting the synthetic "healthy" lung CT images from the "infected" lung CT images. A median filter and K-means clustering were then applied to contour the lesions. The auto segmentation approach was validated on two public datasets (Coronacases and Radiopedia). The Dice coefficients reached 0.748 and 0.730, respectively, for the Coronacases and Radiopedia datasets. Meanwhile, the precision and sensitivity for lesion segmentationdetection are 0.813 and 0.735 for the Coronacases dataset, and 0.773 and 0.726 for the Radiopedia dataset. The performance is comparable to existing supervised segmentation networks and outperforms previous unsupervised ones. The proposed unsupervised segmentation method achieved high accuracy and efficiency in automatic COVID-19 lesion delineation. The segmentation result can serve as a baseline for further manual modification and a quality assurance tool for lesion diagnosis. Furthermore, due to its unsupervised nature, the result is not influenced by physicians' experience which otherwise is crucial for supervised methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge