Bci Competition Datasets
Papers and Code
EEG Foundation Models: Progresses, Benchmarking, and Open Problems
Jan 25, 2026Electroencephalography (EEG) foundation models have recently emerged as a promising paradigm for brain-computer interfaces (BCIs), aiming to learn transferable neural representations from large-scale heterogeneous recordings. Despite rapid progresses, there lacks fair and comprehensive comparisons of existing EEG foundation models, due to inconsistent pre-training objectives, preprocessing choices, and downstream evaluation protocols. This paper fills this gap. We first review 50 representative models and organize their design choices into a unified taxonomic framework including data standardization, model architectures, and self-supervised pre-training strategies. We then evaluate 12 open-source foundation models and competitive specialist baselines across 13 EEG datasets spanning nine BCI paradigms. Emphasizing real-world deployments, we consider both cross-subject generalization under a leave-one-subject-out protocol and rapid calibration under a within-subject few-shot setting. We further compare full-parameter fine-tuning with linear probing to assess the transferability of pre-trained representations, and examine the relationship between model scale and downstream performance. Our results indicate that: 1) linear probing is frequently insufficient; 2) specialist models trained from scratch remain competitive across many tasks; and, 3) larger foundation models do not necessarily yield better generalization performance under current data regimes and training practices.
HCFT: Hierarchical Convolutional Fusion Transformer for EEG Decoding
Jan 18, 2026Electroencephalography (EEG) decoding requires models that can effectively extract and integrate complex temporal, spectral, and spatial features from multichannel signals. To address this challenge, we propose a lightweight and generalizable decoding framework named Hierarchical Convolutional Fusion Transformer (HCFT), which combines dual-branch convolutional encoders and hierarchical Transformer blocks for multi-scale EEG representation learning. Specifically, the model first captures local temporal and spatiotemporal dynamics through time-domain and time-space convolutional branches, and then aligns these features via a cross-attention mechanism that enables interaction between branches at each stage. Subsequently, a hierarchical Transformer fusion structure is employed to encode global dependencies across all feature stages, while a customized Dynamic Tanh normalization module is introduced to replace traditional Layer Normalization in order to enhance training stability and reduce redundancy. Extensive experiments are conducted on two representative benchmark datasets, BCI Competition IV-2b and CHB-MIT, covering both event-related cross-subject classification and continuous seizure prediction tasks. Results show that HCFT achieves 80.83% average accuracy and a Cohen's kappa of 0.6165 on BCI IV-2b, as well as 99.10% sensitivity, 0.0236 false positives per hour, and 98.82% specificity on CHB-MIT, consistently outperforming over ten state-of-the-art baseline methods. Ablation studies confirm that each core component of the proposed framework contributes significantly to the overall decoding performance, demonstrating HCFT's effectiveness in capturing EEG dynamics and its potential for real-world BCI applications.
Meta-cognitive Multi-scale Hierarchical Reasoning for Motor Imagery Decoding
Nov 11, 2025Brain-computer interface (BCI) aims to decode motor intent from noninvasive neural signals to enable control of external devices, but practical deployment remains limited by noise and variability in motor imagery (MI)-based electroencephalogram (EEG) signals. This work investigates a hierarchical and meta-cognitive decoding framework for four-class MI classification. We introduce a multi-scale hierarchical signal processing module that reorganizes backbone features into temporal multi-scale representations, together with an introspective uncertainty estimation module that assigns per-cycle reliability scores and guides iterative refinement. We instantiate this framework on three standard EEG backbones (EEGNet, ShallowConvNet, and DeepConvNet) and evaluate four-class MI decoding using the BCI Competition IV-2a dataset under a subject-independent setting. Across all backbones, the proposed components improve average classification accuracy and reduce inter-subject variance compared to the corresponding baselines, indicating increased robustness to subject heterogeneity and noisy trials. These results suggest that combining hierarchical multi-scale processing with introspective confidence estimation can enhance the reliability of MI-based BCI systems.
Motor Imagery Classification Using Feature Fusion of Spatially Weighted Electroencephalography
Nov 14, 2025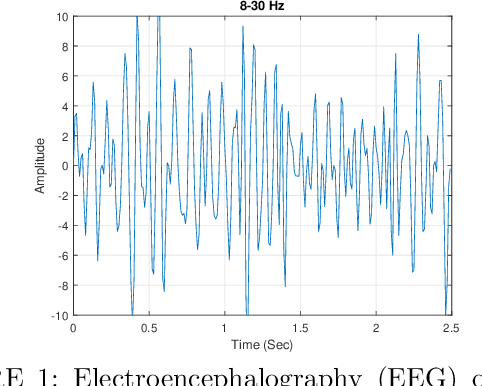
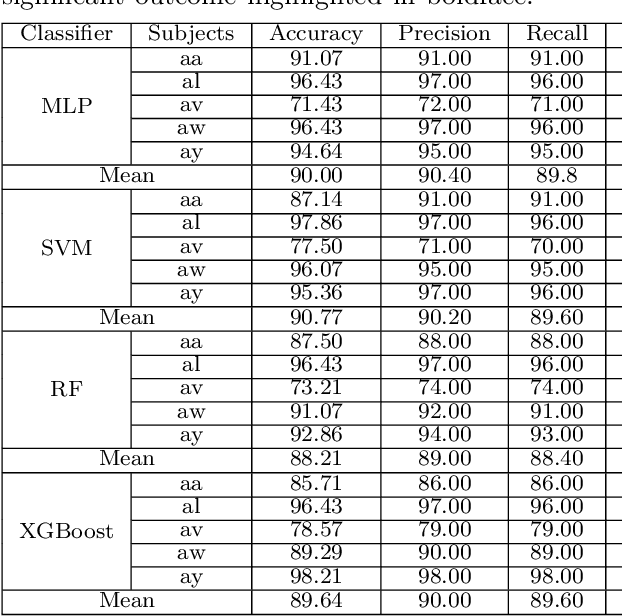
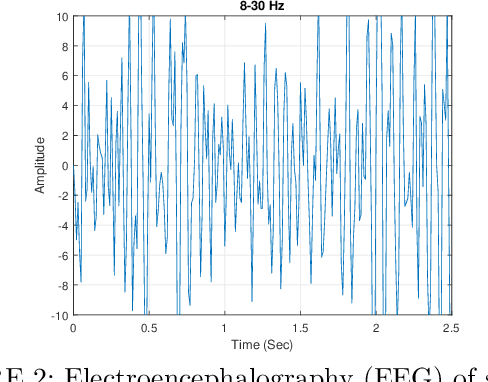
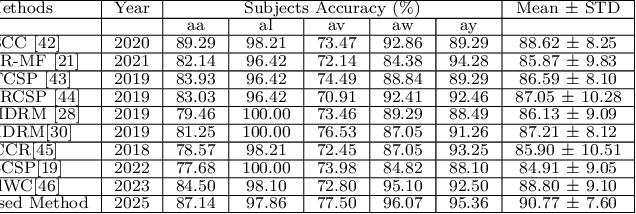
A Brain Computer Interface (BCI) connects the human brain to the outside world, providing a direct communication channel. Electroencephalography (EEG) signals are commonly used in BCIs to reflect cognitive patterns related to motor function activities. However, due to the multichannel nature of EEG signals, explicit information processing is crucial to lessen computational complexity in BCI systems. This study proposes an innovative method based on brain region-specific channel selection and multi-domain feature fusion to improve classification accuracy. The novelty of the proposed approach lies in region-based channel selection, where EEG channels are grouped according to their functional relevance to distinct brain regions. By selecting channels based on specific regions involved in motor imagery (MI) tasks, this technique eliminates irrelevant channels, reducing data dimensionality and improving computational efficiency. This also ensures that the extracted features are more reflective of the brain actual activity related to motor tasks. Three distinct feature extraction methods Common Spatial Pattern (CSP), Fuzzy C-means clustering, and Tangent Space Mapping (TSM), are applied to each group of channels based on their brain region. Each method targets different characteristics of the EEG signal: CSP focuses on spatial patterns, Fuzzy C means identifies clusters within the data, and TSM captures non-linear patterns in the signal. The combined feature vector is used to classify motor imagery tasks (left hand, right hand, and right foot) using Support Vector Machine (SVM). The proposed method was validated on publicly available benchmark EEG datasets (IVA and I) from the BCI competition III and IV. The results show that the approach outperforms existing methods, achieving classification accuracies of 90.77% and 84.50% for datasets IVA and I, respectively.
Biologically Plausible Online Hebbian Meta-Learning: Two-Timescale Local Rules for Spiking Neural Brain Interfaces
Sep 19, 2025



Brain-Computer Interfaces face challenges from neural signal instability and memory constraints for real-time implantable applications. We introduce an online SNN decoder using local three-factor learning rules with dual-timescale eligibility traces that avoid backpropagation through time while maintaining competitive performance. Our approach combines error-modulated Hebbian updates, fast/slow trace consolidation, and adaptive learning rate control, requiring only O(1) memory versus O(T) for BPTT methods. Evaluations on two primate datasets achieve comparable decoding accuracy (Pearson $R \geq 0.63$ Zenodo, $R \geq 0.81$ MC Maze) with 28-35% memory reduction and faster convergence than BPTT-trained SNNs. Closed-loop simulations with synthetic neural populations demonstrate adaptation to neural disruptions and learning from scratch without offline calibration. This work enables memory-efficient, continuously adaptive neural decoding suitable for resource-constrained implantable BCI systems.
TFOC-Net: A Short-time Fourier Transform-based Deep Learning Approach for Enhancing Cross-Subject Motor Imagery Classification
Jul 03, 2025Cross-subject motor imagery (CS-MI) classification in brain-computer interfaces (BCIs) is a challenging task due to the significant variability in Electroencephalography (EEG) patterns across different individuals. This variability often results in lower classification accuracy compared to subject-specific models, presenting a major barrier to developing calibration-free BCIs suitable for real-world applications. In this paper, we introduce a novel approach that significantly enhances cross-subject MI classification performance through optimized preprocessing and deep learning techniques. Our approach involves direct classification of Short-Time Fourier Transform (STFT)-transformed EEG data, optimized STFT parameters, and a balanced batching strategy during training of a Convolutional Neural Network (CNN). This approach is uniquely validated across four different datasets, including three widely-used benchmark datasets leading to substantial improvements in cross-subject classification, achieving 67.60% on the BCI Competition IV Dataset 1 (IV-1), 65.96% on Dataset 2A (IV-2A), and 80.22% on Dataset 2B (IV-2B), outperforming state-of-the-art techniques. Additionally, we systematically investigate the classification performance using MI windows ranging from the full 4-second window to 1-second windows. These results establish a new benchmark for generalizable, calibration-free MI classification in addition to contributing a robust open-access dataset to advance research in this domain.
AGTCNet: A Graph-Temporal Approach for Principled Motor Imagery EEG Classification
Jun 26, 2025Brain-computer interface (BCI) technology utilizing electroencephalography (EEG) marks a transformative innovation, empowering motor-impaired individuals to engage with their environment on equal footing. Despite its promising potential, developing subject-invariant and session-invariant BCI systems remains a significant challenge due to the inherent complexity and variability of neural activity across individuals and over time, compounded by EEG hardware constraints. While prior studies have sought to develop robust BCI systems, existing approaches remain ineffective in capturing the intricate spatiotemporal dependencies within multichannel EEG signals. This study addresses this gap by introducing the attentive graph-temporal convolutional network (AGTCNet), a novel graph-temporal model for motor imagery EEG (MI-EEG) classification. Specifically, AGTCNet leverages the topographic configuration of EEG electrodes as an inductive bias and integrates graph convolutional attention network (GCAT) to jointly learn expressive spatiotemporal EEG representations. The proposed model significantly outperformed existing MI-EEG classifiers, achieving state-of-the-art performance while utilizing a compact architecture, underscoring its effectiveness and practicality for BCI deployment. With a 49.87% reduction in model size, 64.65% faster inference time, and shorter input EEG signal, AGTCNet achieved a moving average accuracy of 66.82% for subject-independent classification on the BCI Competition IV Dataset 2a, which further improved to 82.88% when fine-tuned for subject-specific classification. On the EEG Motor Movement/Imagery Dataset, AGTCNet achieved moving average accuracies of 64.14% and 85.22% for 4-class and 2-class subject-independent classifications, respectively, with further improvements to 72.13% and 90.54% for subject-specific classifications.
MPEC: Manifold-Preserved EEG Classification via an Ensemble of Clustering-Based Classifiers
Apr 30, 2025


Accurate classification of EEG signals is crucial for brain-computer interfaces (BCIs) and neuroprosthetic applications, yet many existing methods fail to account for the non-Euclidean, manifold structure of EEG data, resulting in suboptimal performance. Preserving this manifold information is essential to capture the true geometry of EEG signals, but traditional classification techniques largely overlook this need. To this end, we propose MPEC (Manifold-Preserved EEG Classification via an Ensemble of Clustering-Based Classifiers), that introduces two key innovations: (1) a feature engineering phase that combines covariance matrices and Radial Basis Function (RBF) kernels to capture both linear and non-linear relationships among EEG channels, and (2) a clustering phase that employs a modified K-means algorithm tailored for the Riemannian manifold space, ensuring local geometric sensitivity. Ensembling multiple clustering-based classifiers, MPEC achieves superior results, validated by significant improvements on the BCI Competition IV dataset 2a.
SSTAF: Spatial-Spectral-Temporal Attention Fusion Transformer for Motor Imagery Classification
Apr 17, 2025Brain-computer interfaces (BCI) in electroencephalography (EEG)-based motor imagery classification offer promising solutions in neurorehabilitation and assistive technologies by enabling communication between the brain and external devices. However, the non-stationary nature of EEG signals and significant inter-subject variability cause substantial challenges for developing robust cross-subject classification models. This paper introduces a novel Spatial-Spectral-Temporal Attention Fusion (SSTAF) Transformer specifically designed for upper-limb motor imagery classification. Our architecture consists of a spectral transformer and a spatial transformer, followed by a transformer block and a classifier network. Each module is integrated with attention mechanisms that dynamically attend to the most discriminative patterns across multiple domains, such as spectral frequencies, spatial electrode locations, and temporal dynamics. The short-time Fourier transform is incorporated to extract features in the time-frequency domain to make it easier for the model to obtain a better feature distinction. We evaluated our SSTAF Transformer model on two publicly available datasets, the EEGMMIDB dataset, and BCI Competition IV-2a. SSTAF Transformer achieves an accuracy of 76.83% and 68.30% in the data sets, respectively, outperforms traditional CNN-based architectures and a few existing transformer-based approaches.
ISAM-MTL: Cross-subject multi-task learning model with identifiable spikes and associative memory networks
Jan 30, 2025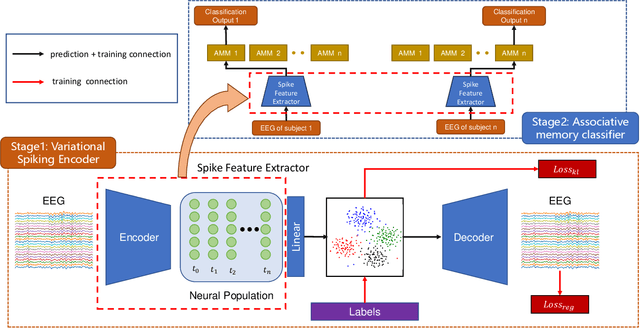
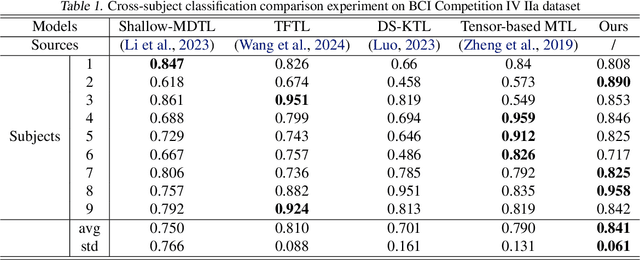
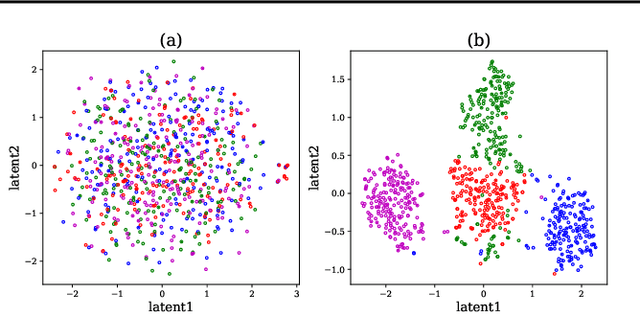
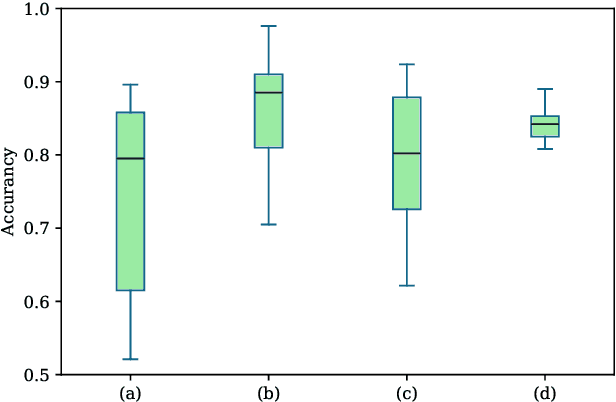
Cross-subject variability in EEG degrades performance of current deep learning models, limiting the development of brain-computer interface (BCI). This paper proposes ISAM-MTL, which is a multi-task learning (MTL) EEG classification model based on identifiable spiking (IS) representations and associative memory (AM) networks. The proposed model treats EEG classification of each subject as an independent task and leverages cross-subject data training to facilitate feature sharing across subjects. ISAM-MTL consists of a spiking feature extractor that captures shared features across subjects and a subject-specific bidirectional associative memory network that is trained by Hebbian learning for efficient and fast within-subject EEG classification. ISAM-MTL integrates learned spiking neural representations with bidirectional associative memory for cross-subject EEG classification. The model employs label-guided variational inference to construct identifiable spike representations, enhancing classification accuracy. Experimental results on two BCI Competition datasets demonstrate that ISAM-MTL improves the average accuracy of cross-subject EEG classification while reducing performance variability among subjects. The model further exhibits the characteristics of few-shot learning and identifiable neural activity beneath EEG, enabling rapid and interpretable calibration for BCI systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge