Jingwei Luo
EEG Foundation Models: Progresses, Benchmarking, and Open Problems
Jan 25, 2026Abstract:Electroencephalography (EEG) foundation models have recently emerged as a promising paradigm for brain-computer interfaces (BCIs), aiming to learn transferable neural representations from large-scale heterogeneous recordings. Despite rapid progresses, there lacks fair and comprehensive comparisons of existing EEG foundation models, due to inconsistent pre-training objectives, preprocessing choices, and downstream evaluation protocols. This paper fills this gap. We first review 50 representative models and organize their design choices into a unified taxonomic framework including data standardization, model architectures, and self-supervised pre-training strategies. We then evaluate 12 open-source foundation models and competitive specialist baselines across 13 EEG datasets spanning nine BCI paradigms. Emphasizing real-world deployments, we consider both cross-subject generalization under a leave-one-subject-out protocol and rapid calibration under a within-subject few-shot setting. We further compare full-parameter fine-tuning with linear probing to assess the transferability of pre-trained representations, and examine the relationship between model scale and downstream performance. Our results indicate that: 1) linear probing is frequently insufficient; 2) specialist models trained from scratch remain competitive across many tasks; and, 3) larger foundation models do not necessarily yield better generalization performance under current data regimes and training practices.
MIRepNet: A Pipeline and Foundation Model for EEG-Based Motor Imagery Classification
Jul 27, 2025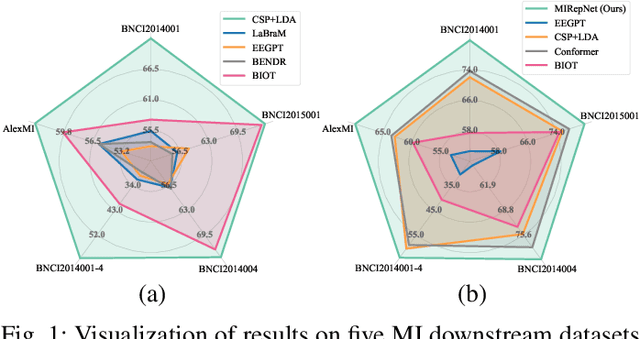
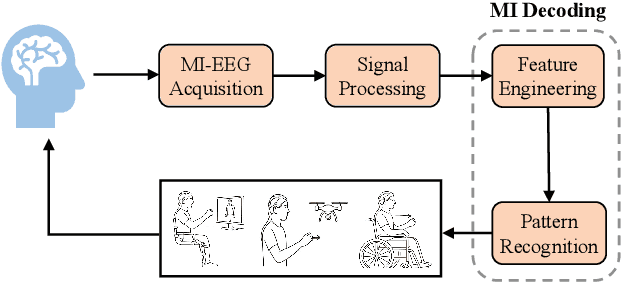
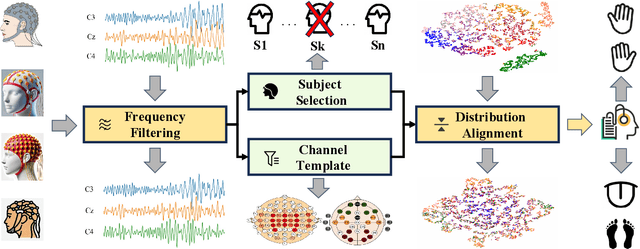
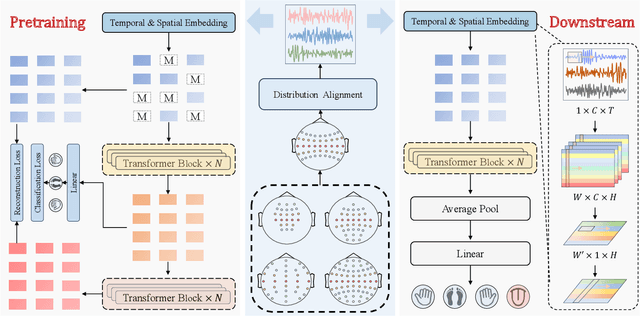
Abstract:Brain-computer interfaces (BCIs) enable direct communication between the brain and external devices. Recent EEG foundation models aim to learn generalized representations across diverse BCI paradigms. However, these approaches overlook fundamental paradigm-specific neurophysiological distinctions, limiting their generalization ability. Importantly, in practical BCI deployments, the specific paradigm such as motor imagery (MI) for stroke rehabilitation or assistive robotics, is generally determined prior to data acquisition. This paper proposes MIRepNet, the first EEG foundation model tailored for the MI paradigm. MIRepNet comprises a high-quality EEG preprocessing pipeline incorporating a neurophysiologically-informed channel template, adaptable to EEG headsets with arbitrary electrode configurations. Furthermore, we introduce a hybrid pretraining strategy that combines self-supervised masked token reconstruction and supervised MI classification, facilitating rapid adaptation and accurate decoding on novel downstream MI tasks with fewer than 30 trials per class. Extensive evaluations across five public MI datasets demonstrated that MIRepNet consistently achieved state-of-the-art performance, significantly outperforming both specialized and generalized EEG models. Our code will be available on GitHub\footnote{https://github.com/staraink/MIRepNet}.
Channel Reflection: Knowledge-Driven Data Augmentation for EEG-Based Brain-Computer Interfaces
Dec 04, 2024Abstract:A brain-computer interface (BCI) enables direct communication between the human brain and external devices. Electroencephalography (EEG) based BCIs are currently the most popular for able-bodied users. To increase user-friendliness, usually a small amount of user-specific EEG data are used for calibration, which may not be enough to develop a pure data-driven decoding model. To cope with this typical calibration data shortage challenge in EEG-based BCIs, this paper proposes a parameter-free channel reflection (CR) data augmentation approach that incorporates prior knowledge on the channel distributions of different BCI paradigms in data augmentation. Experiments on eight public EEG datasets across four different BCI paradigms (motor imagery, steady-state visual evoked potential, P300, and seizure classifications) using different decoding algorithms demonstrated that: 1) CR is effective, i.e., it can noticeably improve the classification accuracy; 2) CR is robust, i.e., it consistently outperforms existing data augmentation approaches in the literature; and, 3) CR is flexible, i.e., it can be combined with other data augmentation approaches to further increase the performance. We suggest that data augmentation approaches like CR should be an essential step in EEG-based BCIs. Our code is available online.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge