Zhixiong Zhang
UniReason 1.0: A Unified Reasoning Framework for World Knowledge Aligned Image Generation and Editing
Feb 02, 2026Abstract:Unified multimodal models often struggle with complex synthesis tasks that demand deep reasoning, and typically treat text-to-image generation and image editing as isolated capabilities rather than interconnected reasoning steps. To address this, we propose UniReason, a unified framework that harmonizes these two tasks through a dual reasoning paradigm. We formulate generation as world knowledge-enhanced planning to inject implicit constraints, and leverage editing capabilities for fine-grained visual refinement to further correct visual errors via self-reflection. This approach unifies generation and editing within a shared representation, mirroring the human cognitive process of planning followed by refinement. We support this framework by systematically constructing a large-scale reasoning-centric dataset (~300k samples) covering five major knowledge domains (e.g., cultural commonsense, physics, etc.) for planning, alongside an agent-generated corpus for visual self-correction. Extensive experiments demonstrate that UniReason achieves advanced performance on reasoning-intensive benchmarks such as WISE, KrisBench and UniREditBench, while maintaining superior general synthesis capabilities.
CODA: Coordinating the Cerebrum and Cerebellum for a Dual-Brain Computer Use Agent with Decoupled Reinforcement Learning
Aug 27, 2025Abstract:Autonomous agents for Graphical User Interfaces (GUIs) face significant challenges in specialized domains such as scientific computing, where both long-horizon planning and precise execution are required. Existing approaches suffer from a trade-off: generalist agents excel at planning but perform poorly in execution, while specialized agents demonstrate the opposite weakness. Recent compositional frameworks attempt to bridge this gap by combining a planner and an actor, but they are typically static and non-trainable, which prevents adaptation from experience. This is a critical limitation given the scarcity of high-quality data in scientific domains. To address these limitations, we introduce CODA, a novel and trainable compositional framework that integrates a generalist planner (Cerebrum) with a specialist executor (Cerebellum), trained via a dedicated two-stage pipeline. In the first stage, Specialization, we apply a decoupled GRPO approach to train an expert planner for each scientific application individually, bootstrapping from a small set of task trajectories. In the second stage, Generalization, we aggregate all successful trajectories from the specialized experts to build a consolidated dataset, which is then used for supervised fine-tuning of the final planner. This equips CODA with both robust execution and cross-domain generalization. Evaluated on four challenging applications from the ScienceBoard benchmark, CODA significantly outperforms baselines and establishes a new state of the art among open-source models.
DivIL: Unveiling and Addressing Over-Invariance for Out-of- Distribution Generalization
Feb 18, 2025Abstract:Out-of-distribution generalization is a common problem that expects the model to perform well in the different distributions even far from the train data. A popular approach to addressing this issue is invariant learning (IL), in which the model is compiled to focus on invariant features instead of spurious features by adding strong constraints during training. However, there are some potential pitfalls of strong invariant constraints. Due to the limited number of diverse environments and over-regularization in the feature space, it may lead to a loss of important details in the invariant features while alleviating the spurious correlations, namely the over-invariance, which can also degrade the generalization performance. We theoretically define the over-invariance and observe that this issue occurs in various classic IL methods. To alleviate this issue, we propose a simple approach Diverse Invariant Learning (DivIL) by adding the unsupervised contrastive learning and the random masking mechanism compensatory for the invariant constraints, which can be applied to various IL methods. Furthermore, we conduct experiments across multiple modalities across 12 datasets and 6 classic models, verifying our over-invariance insight and the effectiveness of our DivIL framework. Our code is available at https://github.com/kokolerk/DivIL.
SongGen: A Single Stage Auto-regressive Transformer for Text-to-Song Generation
Feb 18, 2025Abstract:Text-to-song generation, the task of creating vocals and accompaniment from textual inputs, poses significant challenges due to domain complexity and data scarcity. Existing approaches often employ multi-stage generation procedures, resulting in cumbersome training and inference pipelines. In this paper, we propose SongGen, a fully open-source, single-stage auto-regressive transformer designed for controllable song generation. The proposed model facilitates fine-grained control over diverse musical attributes, including lyrics and textual descriptions of instrumentation, genre, mood, and timbre, while also offering an optional three-second reference clip for voice cloning. Within a unified auto-regressive framework, SongGen supports two output modes: mixed mode, which generates a mixture of vocals and accompaniment directly, and dual-track mode, which synthesizes them separately for greater flexibility in downstream applications. We explore diverse token pattern strategies for each mode, leading to notable improvements and valuable insights. Furthermore, we design an automated data preprocessing pipeline with effective quality control. To foster community engagement and future research, we will release our model weights, training code, annotated data, and preprocessing pipeline. The generated samples are showcased on our project page at https://liuzh-19.github.io/SongGen/ , and the code will be available at https://github.com/LiuZH-19/SongGen .
GPT4Scene: Understand 3D Scenes from Videos with Vision-Language Models
Jan 03, 2025



Abstract:In recent years, 2D Vision-Language Models (VLMs) have made significant strides in image-text understanding tasks. However, their performance in 3D spatial comprehension, which is critical for embodied intelligence, remains limited. Recent advances have leveraged 3D point clouds and multi-view images as inputs, yielding promising results. However, we propose exploring a purely vision-based solution inspired by human perception, which merely relies on visual cues for 3D spatial understanding. This paper empirically investigates the limitations of VLMs in 3D spatial knowledge, revealing that their primary shortcoming lies in the lack of global-local correspondence between the scene and individual frames. To address this, we introduce GPT4Scene, a novel visual prompting paradigm in VLM training and inference that helps build the global-local relationship, significantly improving the 3D spatial understanding of indoor scenes. Specifically, GPT4Scene constructs a 3D Bird's Eye View (BEV) image from the video and marks consistent object IDs across both frames and the BEV image. The model then inputs the concatenated BEV image and video frames with markers. In zero-shot evaluations, GPT4Scene improves performance over closed-source VLMs like GPT-4o. Additionally, we prepare a processed video dataset consisting of 165K text annotation to fine-tune open-source VLMs, achieving state-of-the-art performance on all 3D understanding tasks. Surprisingly, after training with the GPT4Scene paradigm, VLMs consistently improve during inference, even without visual prompting and BEV image as explicit correspondence. It demonstrates that the proposed paradigm helps VLMs develop an intrinsic ability to understand 3D scenes, which paves the way for a noninvasive approach to extending pre-trained VLMs for 3D scene understanding.
SparkRA: A Retrieval-Augmented Knowledge Service System Based on Spark Large Language Model
Aug 13, 2024
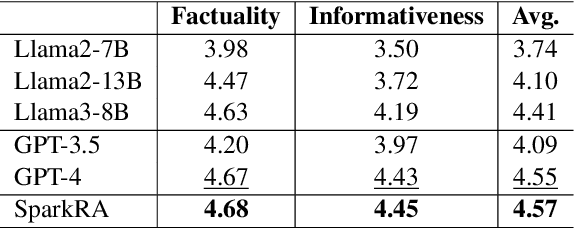

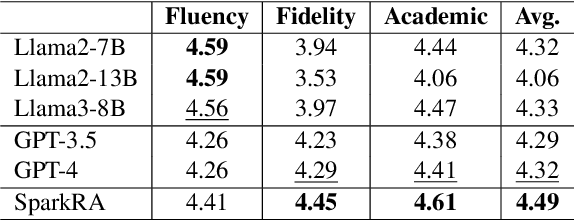
Abstract:Large language models (LLMs) have shown remarkable achievements across various language tasks.To enhance the performance of LLMs in scientific literature services, we developed the scientific literature LLM (SciLit-LLM) through pre-training and supervised fine-tuning on scientific literature, building upon the iFLYTEK Spark LLM. Furthermore, we present a knowledge service system Spark Research Assistant (SparkRA) based on our SciLit-LLM. SparkRA is accessible online and provides three primary functions: literature investigation, paper reading, and academic writing. As of July 30, 2024, SparkRA has garnered over 50,000 registered users, with a total usage count exceeding 1.3 million.
Partial Annotation Learning for Biomedical Entity Recognition
May 22, 2023Abstract:Motivation: Named Entity Recognition (NER) is a key task to support biomedical research. In Biomedical Named Entity Recognition (BioNER), obtaining high-quality expert annotated data is laborious and expensive, leading to the development of automatic approaches such as distant supervision. However, manually and automatically generated data often suffer from the unlabeled entity problem, whereby many entity annotations are missing, degrading the performance of full annotation NER models. Results: To address this problem, we systematically study the effectiveness of partial annotation learning methods for biomedical entity recognition over different simulated scenarios of missing entity annotations. Furthermore, we propose a TS-PubMedBERT-Partial-CRF partial annotation learning model. We harmonize 15 biomedical NER corpora encompassing five entity types to serve as a gold standard and compare against two commonly used partial annotation learning models, BiLSTM-Partial-CRF and EER-PubMedBERT, and the state-of-the-art full annotation learning BioNER model PubMedBERT tagger. Results show that partial annotation learning-based methods can effectively learn from biomedical corpora with missing entity annotations. Our proposed model outperforms alternatives and, specifically, the PubMedBERT tagger by 38% in F1-score under high missing entity rates. The recall of entity mentions in our model is also competitive with the upper bound on the fully annotated dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge