Yuqian Zhao
Towards a Single Unified Model for Effective Detection, Segmentation, and Diagnosis of Eight Major Cancers Using a Large Collection of CT Scans
Jan 28, 2023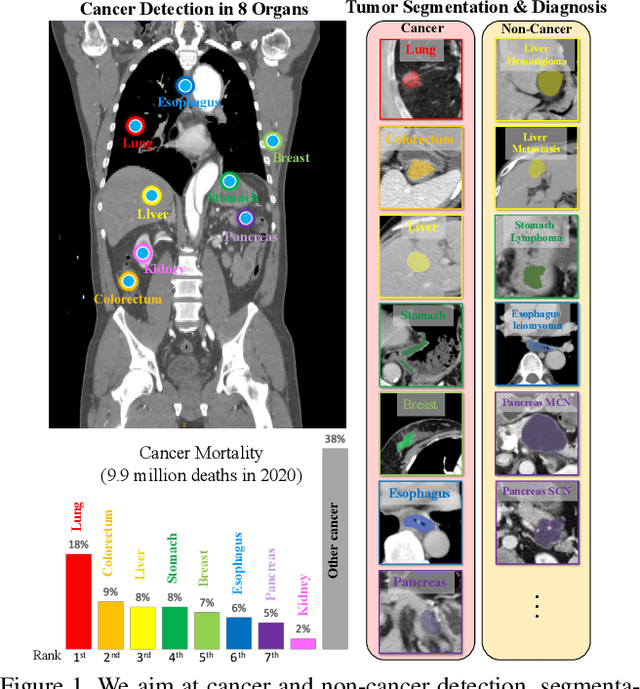
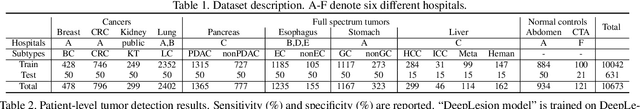
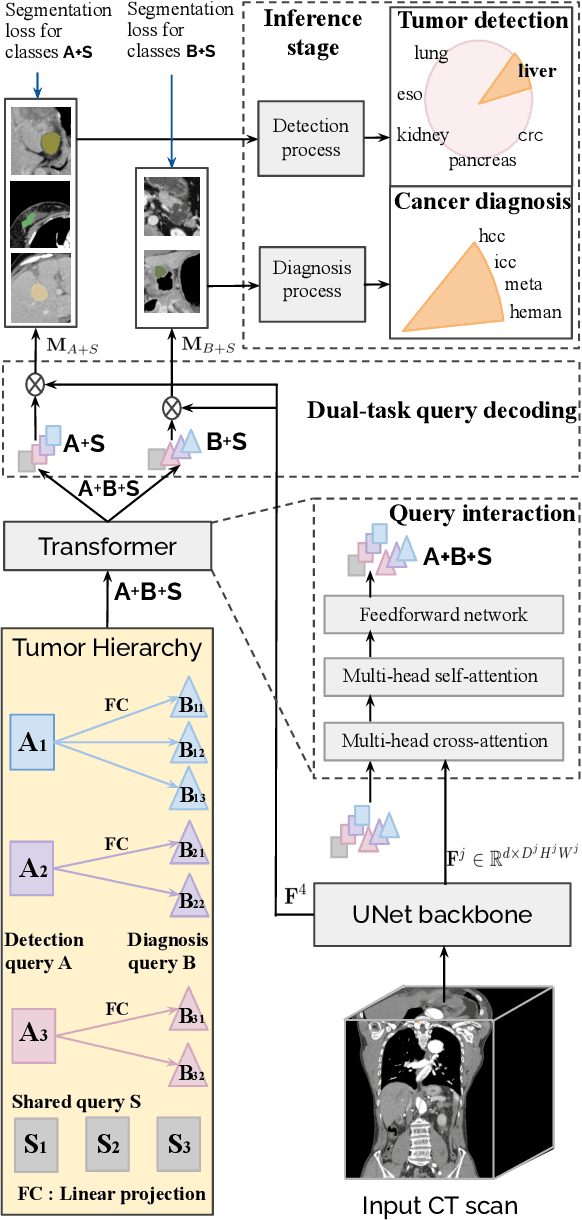
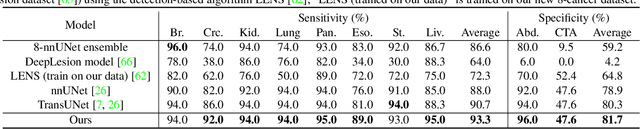
Abstract:Human readers or radiologists routinely perform full-body multi-organ multi-disease detection and diagnosis in clinical practice, while most medical AI systems are built to focus on single organs with a narrow list of a few diseases. This might severely limit AI's clinical adoption. A certain number of AI models need to be assembled non-trivially to match the diagnostic process of a human reading a CT scan. In this paper, we construct a Unified Tumor Transformer (UniT) model to detect (tumor existence and location) and diagnose (tumor characteristics) eight major cancer-prevalent organs in CT scans. UniT is a query-based Mask Transformer model with the output of multi-organ and multi-tumor semantic segmentation. We decouple the object queries into organ queries, detection queries and diagnosis queries, and further establish hierarchical relationships among the three groups. This clinically-inspired architecture effectively assists inter- and intra-organ representation learning of tumors and facilitates the resolution of these complex, anatomically related multi-organ cancer image reading tasks. UniT is trained end-to-end using a curated large-scale CT images of 10,042 patients including eight major types of cancers and occurring non-cancer tumors (all are pathology-confirmed with 3D tumor masks annotated by radiologists). On the test set of 631 patients, UniT has demonstrated strong performance under a set of clinically relevant evaluation metrics, substantially outperforming both multi-organ segmentation methods and an assembly of eight single-organ expert models in tumor detection, segmentation, and diagnosis. Such a unified multi-cancer image reading model (UniT) can significantly reduce the number of false positives produced by combined multi-system models. This moves one step closer towards a universal high-performance cancer screening tool.
IMKGA-SM: Interpretable Multimodal Knowledge Graph Answer Prediction via Sequence Modeling
Jan 11, 2023



Abstract:Multimodal knowledge graph link prediction aims to improve the accuracy and efficiency of link prediction tasks for multimodal data. However, for complex multimodal information and sparse training data, it is usually difficult to achieve interpretability and high accuracy simultaneously for most methods. To address this difficulty, a new model is developed in this paper, namely Interpretable Multimodal Knowledge Graph Answer Prediction via Sequence Modeling (IMKGA-SM). First, a multi-modal fine-grained fusion method is proposed, and Vgg16 and Optical Character Recognition (OCR) techniques are adopted to effectively extract text information from images and images. Then, the knowledge graph link prediction task is modelled as an offline reinforcement learning Markov decision model, which is then abstracted into a unified sequence framework. An interactive perception-based reward expectation mechanism and a special causal masking mechanism are designed, which "converts" the query into an inference path. Then, an autoregressive dynamic gradient adjustment mechanism is proposed to alleviate the insufficient problem of multimodal optimization. Finally, two datasets are adopted for experiments, and the popular SOTA baselines are used for comparison. The results show that the developed IMKGA-SM achieves much better performance than SOTA baselines on multimodal link prediction datasets of different sizes.
Knowledge-aware Deep Framework for Collaborative Skin Lesion Segmentation and Melanoma Recognition
Jun 07, 2021



Abstract:Deep learning techniques have shown their superior performance in dermatologist clinical inspection. Nevertheless, melanoma diagnosis is still a challenging task due to the difficulty of incorporating the useful dermatologist clinical knowledge into the learning process. In this paper, we propose a novel knowledge-aware deep framework that incorporates some clinical knowledge into collaborative learning of two important melanoma diagnosis tasks, i.e., skin lesion segmentation and melanoma recognition. Specifically, to exploit the knowledge of morphological expressions of the lesion region and also the periphery region for melanoma identification, a lesion-based pooling and shape extraction (LPSE) scheme is designed, which transfers the structure information obtained from skin lesion segmentation into melanoma recognition. Meanwhile, to pass the skin lesion diagnosis knowledge from melanoma recognition to skin lesion segmentation, an effective diagnosis guided feature fusion (DGFF) strategy is designed. Moreover, we propose a recursive mutual learning mechanism that further promotes the inter-task cooperation, and thus iteratively improves the joint learning capability of the model for both skin lesion segmentation and melanoma recognition. Experimental results on two publicly available skin lesion datasets show the effectiveness of the proposed method for melanoma analysis.
Efficient Misalignment-Robust Multi-Focus Microscopical Images Fusion
Dec 21, 2018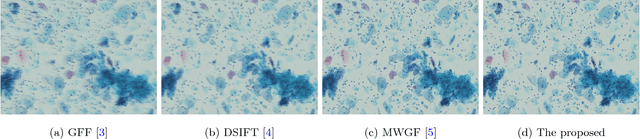
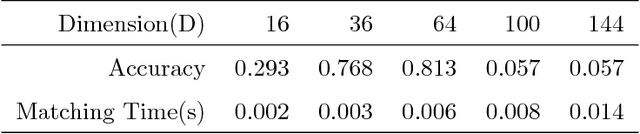
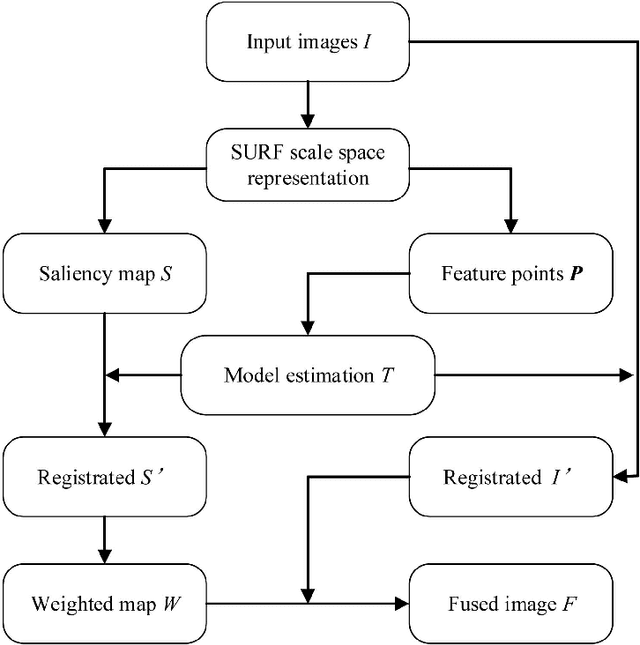

Abstract:In this paper we propose a very efficient method to fuse the unregistered multi-focus microscopical images based on the speed-up robust features (SURF). Our method follows the pipeline of first registration and then fusion. However, instead of treating the registration and fusion as two completely independent stage, we propose to reuse the determinant of the approximate Hessian generated in SURF detection stage as the corresponding salient response for the final image fusion, thus it enables nearly cost-free saliency map generation. In addition, due to the adoption of SURF scale space representation, our method can generate scale-invariant saliency map which is desired for scale-invariant image fusion. We present an extensive evaluation on the dataset consisting of several groups of unregistered multi-focus 4K ultra HD microscopic images with size of 4112 x 3008. Compared with the state-of-the-art multi-focus image fusion methods, our method is much faster and achieve better results in the visual performance. Our method provides a flexible and efficient way to integrate complementary and redundant information from multiple multi-focus ultra HD unregistered images into a fused image that contains better description than any of the individual input images. Code is available at https://github.com/yiqingmy/JointRF.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge