Yongshuai Hou
Evaluating and Steering Modality Preferences in Multimodal Large Language Model
May 27, 2025Abstract:Multimodal large language models (MLLMs) have achieved remarkable performance on complex tasks with multimodal context. However, it is still understudied whether they exhibit modality preference when processing multimodal contexts. To study this question, we first build a \textbf{MC\textsuperscript{2}} benchmark under controlled evidence conflict scenarios to systematically evaluate modality preference, which is the tendency to favor one modality over another when making decisions based on multimodal conflicting evidence. Our extensive evaluation reveals that all 18 tested MLLMs generally demonstrate clear modality bias, and modality preference can be influenced by external interventions. An in-depth analysis reveals that the preference direction can be captured within the latent representations of MLLMs. Built on this, we propose a probing and steering method based on representation engineering to explicitly control modality preference without additional fine-tuning or carefully crafted prompts. Our method effectively amplifies modality preference toward a desired direction and applies to downstream tasks such as hallucination mitigation and multimodal machine translation, yielding promising improvements.
SHAPE: A Sample-adaptive Hierarchical Prediction Network for Medication Recommendation
Sep 09, 2023



Abstract:Effectively medication recommendation with complex multimorbidity conditions is a critical task in healthcare. Most existing works predicted medications based on longitudinal records, which assumed the information transmitted patterns of learning longitudinal sequence data are stable and intra-visit medical events are serialized. However, the following conditions may have been ignored: 1) A more compact encoder for intra-relationship in the intra-visit medical event is urgent; 2) Strategies for learning accurate representations of the variable longitudinal sequences of patients are different. In this paper, we proposed a novel Sample-adaptive Hierarchical medicAtion Prediction nEtwork, termed SHAPE, to tackle the above challenges in the medication recommendation task. Specifically, we design a compact intra-visit set encoder to encode the relationship in the medical event for obtaining visit-level representation and then develop an inter-visit longitudinal encoder to learn the patient-level longitudinal representation efficiently. To endow the model with the capability of modeling the variable visit length, we introduce a soft curriculum learning method to assign the difficulty of each sample automatically by the visit length. Extensive experiments on a benchmark dataset verify the superiority of our model compared with several state-of-the-art baselines.
Nebula-I: A General Framework for Collaboratively Training Deep Learning Models on Low-Bandwidth Cloud Clusters
May 19, 2022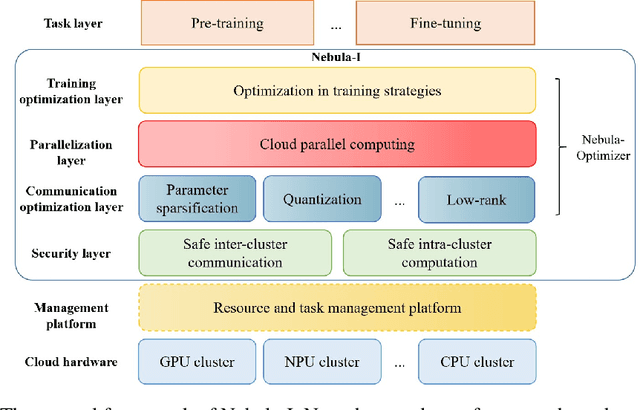
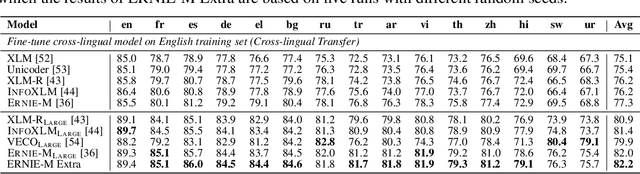
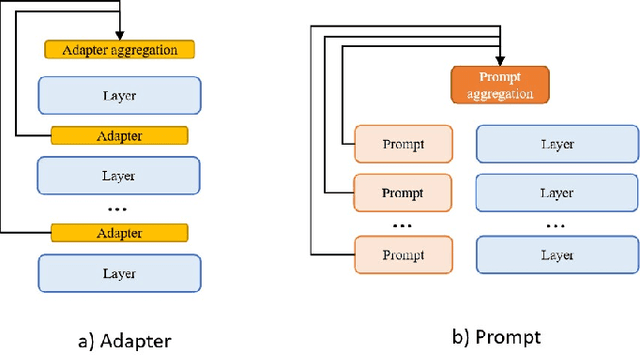
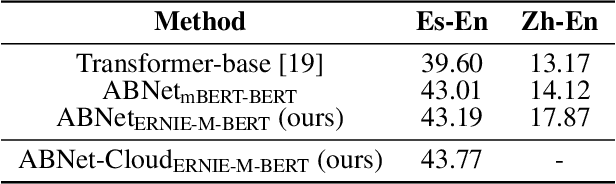
Abstract:The ever-growing model size and scale of compute have attracted increasing interests in training deep learning models over multiple nodes. However, when it comes to training on cloud clusters, especially across remote clusters, huge challenges are faced. In this work, we introduce a general framework, Nebula-I, for collaboratively training deep learning models over remote heterogeneous clusters, the connections between which are low-bandwidth wide area networks (WANs). We took natural language processing (NLP) as an example to show how Nebula-I works in different training phases that include: a) pre-training a multilingual language model using two remote clusters; and b) fine-tuning a machine translation model using knowledge distilled from pre-trained models, which run through the most popular paradigm of recent deep learning. To balance the accuracy and communication efficiency, in Nebula-I, parameter-efficient training strategies, hybrid parallel computing methods and adaptive communication acceleration techniques are jointly applied. Meanwhile, security strategies are employed to guarantee the safety, reliability and privacy in intra-cluster computation and inter-cluster communication. Nebula-I is implemented with the PaddlePaddle deep learning framework, which can support collaborative training over heterogeneous hardware, e.g. GPU and NPU. Experiments demonstrate that the proposed framework could substantially maximize the training efficiency while preserving satisfactory NLP performance. By using Nebula-I, users can run large-scale training tasks over cloud clusters with minimum developments, and the utility of existed large pre-trained models could be further promoted. We also introduced new state-of-the-art results on cross-lingual natural language inference tasks, which are generated based upon a novel learning framework and Nebula-I.
Two heads are better than one: Enhancing medical representations by pre-training over structured and unstructured electronic health records
Jan 25, 2022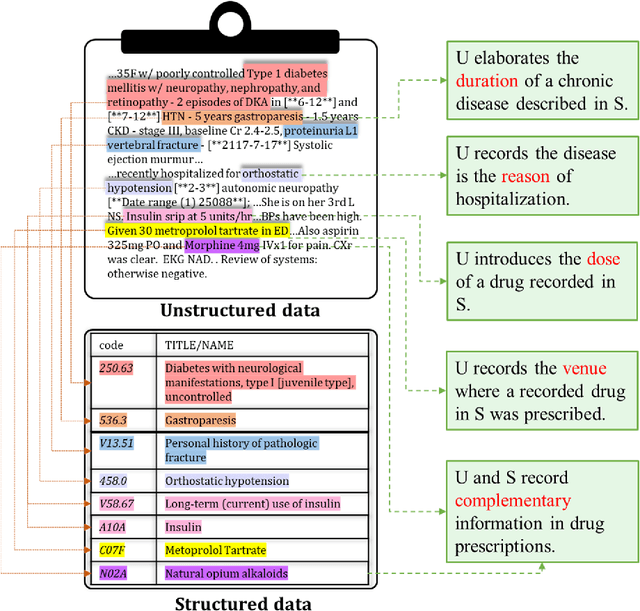
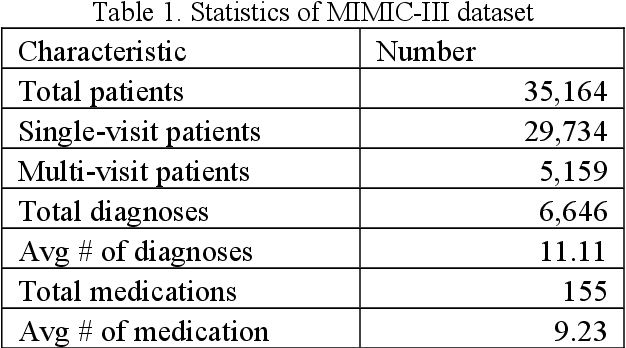
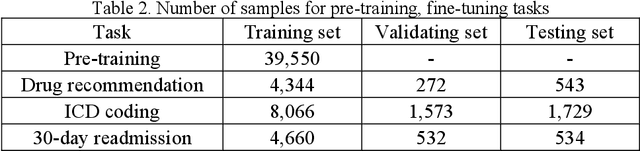
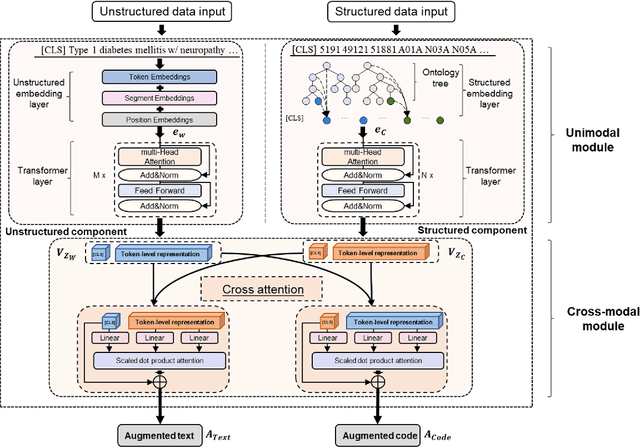
Abstract:The massive context of electronic health records (EHRs) has created enormous potentials for improving healthcare, among which structured (coded) data and unstructured (text) data are two important textual modalities. They do not exist in isolation and can complement each other in most real-life clinical scenarios. Most existing researches in medical informatics, however, either only focus on a particular modality or straightforwardly concatenate the information from different modalities, which ignore the interaction and information sharing between them. To address these issues, we proposed a unified deep learning-based medical pre-trained language model, named UMM-PLM, to automatically learn representative features from multimodal EHRs that consist of both structured data and unstructured data. Specifically, we first developed parallel unimodal information representation modules to capture the unimodal-specific characteristic, where unimodal representations were learned from each data source separately. A cross-modal module was further introduced to model the interactions between different modalities. We pre-trained the model on a large EHRs dataset containing both structured data and unstructured data and verified the effectiveness of the model on three downstream clinical tasks, i.e., medication recommendation, 30-day readmission and ICD coding through extensive experiments. The results demonstrate the power of UMM-PLM compared with benchmark methods and state-of-the-art baselines. Analyses show that UMM-PLM can effectively concern with multimodal textual information and has the potential to provide more comprehensive interpretations for clinical decision making.
Machine Learning for Multimodal Electronic Health Records-based Research: Challenges and Perspectives
Nov 09, 2021



Abstract:Background: Electronic Health Records (EHRs) contain rich information of patients' health history, which usually include both structured and unstructured data. There have been many studies focusing on distilling valuable information from structured data, such as disease codes, laboratory test results, and treatments. However, relying on structured data only might be insufficient in reflecting patients' comprehensive information and such data may occasionally contain erroneous records. Objective: With the recent advances of machine learning (ML) and deep learning (DL) techniques, an increasing number of studies seek to obtain more accurate results by incorporating unstructured free-text data as well. This paper reviews studies that use multimodal data, i.e. a combination of structured and unstructured data, from EHRs as input for conventional ML or DL models to address the targeted tasks. Materials and Methods: We searched in the Institute of Electrical and Electronics Engineers (IEEE) Digital Library, PubMed, and Association for Computing Machinery (ACM) Digital Library for articles related to ML-based multimodal EHR studies. Results and Discussion: With the final 94 included studies, we focus on how data from different modalities were combined and interacted using conventional ML and DL techniques, and how these algorithms were applied in EHR-related tasks. Further, we investigate the advantages and limitations of these fusion methods and indicate future directions for ML-based multimodal EHR research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge