Sicen Liu
Fairness Is Not Just Ethical: Performance Trade-Off via Data Correlation Tuning to Mitigate Bias in ML Software
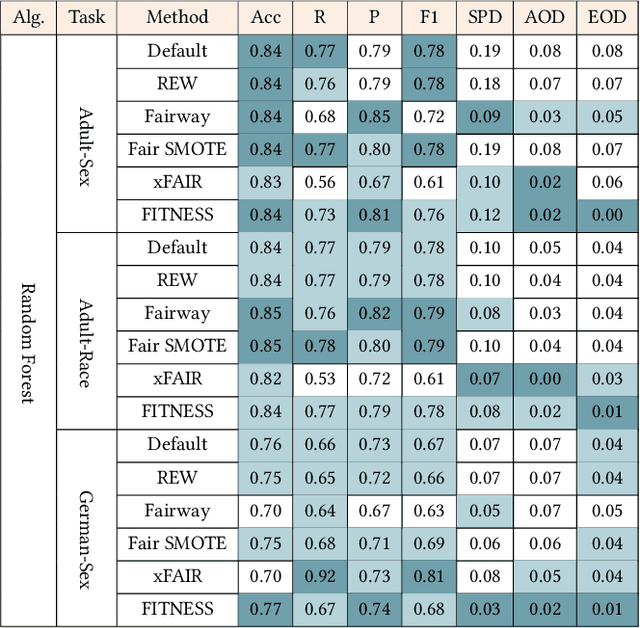
Dec 19, 2025Abstract:Traditional software fairness research typically emphasizes ethical and social imperatives, neglecting that fairness fundamentally represents a core software quality issue arising directly from performance disparities across sensitive user groups. Recognizing fairness explicitly as a software quality dimension yields practical benefits beyond ethical considerations, notably improved predictive performance for unprivileged groups, enhanced out-of-distribution generalization, and increased geographic transferability in real-world deployments. Nevertheless, existing bias mitigation methods face a critical dilemma: while pre-processing methods offer broad applicability across model types, they generally fall short in effectiveness compared to post-processing techniques. To overcome this challenge, we propose Correlation Tuning (CoT), a novel pre-processing approach designed to mitigate bias by adjusting data correlations. Specifically, CoT introduces the Phi-coefficient, an intuitive correlation measure, to systematically quantify correlation between sensitive attributes and labels, and employs multi-objective optimization to address the proxy biases. Extensive evaluations demonstrate that CoT increases the true positive rate of unprivileged groups by an average of 17.5% and reduces three key bias metrics, including statistical parity difference (SPD), average odds difference (AOD), and equal opportunity difference (EOD), by more than 50% on average. CoT outperforms state-of-the-art methods by three and ten percentage points in single attribute and multiple attributes scenarios, respectively. We will publicly release our experimental results and source code to facilitate future research.
Self-eXplainable AI for Medical Image Analysis: A Survey and New Outlooks
Oct 03, 2024



Abstract:The increasing demand for transparent and reliable models, particularly in high-stakes decision-making areas such as medical image analysis, has led to the emergence of eXplainable Artificial Intelligence (XAI). Post-hoc XAI techniques, which aim to explain black-box models after training, have been controversial in recent works concerning their fidelity to the models' predictions. In contrast, Self-eXplainable AI (S-XAI) offers a compelling alternative by incorporating explainability directly into the training process of deep learning models. This approach allows models to generate inherent explanations that are closely aligned with their internal decision-making processes. Such enhanced transparency significantly supports the trustworthiness, robustness, and accountability of AI systems in real-world medical applications. To facilitate the development of S-XAI methods for medical image analysis, this survey presents an comprehensive review across various image modalities and clinical applications. It covers more than 200 papers from three key perspectives: 1) input explainability through the integration of explainable feature engineering and knowledge graph, 2) model explainability via attention-based learning, concept-based learning, and prototype-based learning, and 3) output explainability by providing counterfactual explanation and textual explanation. Additionally, this paper outlines the desired characteristics of explainability and existing evaluation methods for assessing explanation quality. Finally, it discusses the major challenges and future research directions in developing S-XAI for medical image analysis.
SHAPE: A Sample-adaptive Hierarchical Prediction Network for Medication Recommendation
Sep 09, 2023



Abstract:Effectively medication recommendation with complex multimorbidity conditions is a critical task in healthcare. Most existing works predicted medications based on longitudinal records, which assumed the information transmitted patterns of learning longitudinal sequence data are stable and intra-visit medical events are serialized. However, the following conditions may have been ignored: 1) A more compact encoder for intra-relationship in the intra-visit medical event is urgent; 2) Strategies for learning accurate representations of the variable longitudinal sequences of patients are different. In this paper, we proposed a novel Sample-adaptive Hierarchical medicAtion Prediction nEtwork, termed SHAPE, to tackle the above challenges in the medication recommendation task. Specifically, we design a compact intra-visit set encoder to encode the relationship in the medical event for obtaining visit-level representation and then develop an inter-visit longitudinal encoder to learn the patient-level longitudinal representation efficiently. To endow the model with the capability of modeling the variable visit length, we introduce a soft curriculum learning method to assign the difficulty of each sample automatically by the visit length. Extensive experiments on a benchmark dataset verify the superiority of our model compared with several state-of-the-art baselines.
Medication Recommendation via Domain Knowledge Informed Deep Learning
May 31, 2023



Abstract:Medication recommendation is a fundamental yet crucial branch of healthcare, which provides opportunities to support clinical physicians with more accurate medication prescriptions for patients with complex health conditions. Learning from electronic health records (EHR) to recommend medications is the most common way in previous studies. However, most of them neglect incorporating domain knowledge according to the clinical manifestations in the EHR of the patient. To address these issues, we propose a novel \textbf{D}omain \textbf{K}nowledge \textbf{I}nformed \textbf{Net}work (DKINet) to integrate domain knowledge with observable clinical manifestations of the patient, which is the first dynamic domain knowledge informed framework toward medication recommendation. In particular, we first design a knowledge-driven encoder to capture the domain information and then develop a data-driven encoder to integrate domain knowledge into the observable EHR. To endow the model with the capability of temporal decision, we design an explicit medication encoder for learning the longitudinal dependence of the patient. Extensive experiments on three publicly available datasets verify the superiority of our method. The code will be public upon acceptance.
FITNESS: A Causal De-correlation Approach for Mitigating Bias in Machine Learning Software
May 23, 2023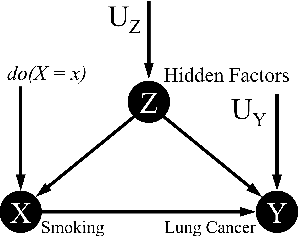

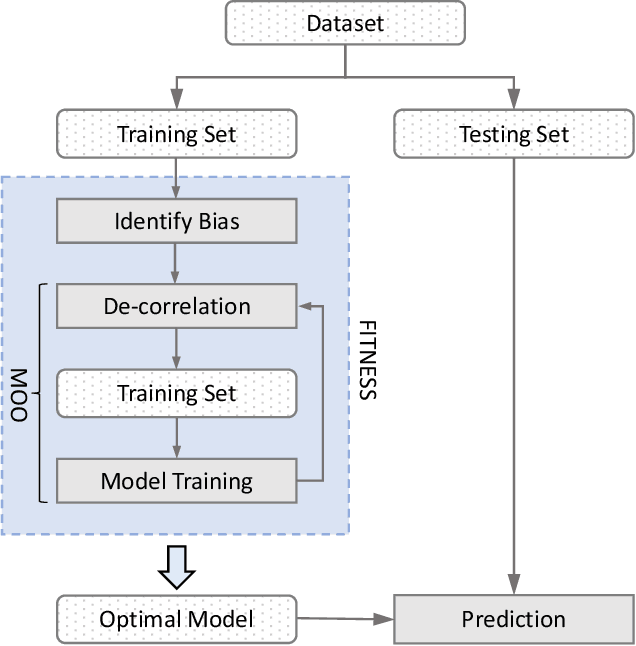
Abstract:Software built on top of machine learning algorithms is becoming increasingly prevalent in a variety of fields, including college admissions, healthcare, insurance, and justice. The effectiveness and efficiency of these systems heavily depend on the quality of the training datasets. Biased datasets can lead to unfair and potentially harmful outcomes, particularly in such critical decision-making systems where the allocation of resources may be affected. This can exacerbate discrimination against certain groups and cause significant social disruption. To mitigate such unfairness, a series of bias-mitigating methods are proposed. Generally, these studies improve the fairness of the trained models to a certain degree but with the expense of sacrificing the model performance. In this paper, we propose FITNESS, a bias mitigation approach via de-correlating the causal effects between sensitive features (e.g., the sex) and the label. Our key idea is that by de-correlating such effects from a causality perspective, the model would avoid making predictions based on sensitive features and thus fairness could be improved. Furthermore, FITNESS leverages multi-objective optimization to achieve a better performance-fairness trade-off. To evaluate the effectiveness, we compare FITNESS with 7 state-of-the-art methods in 8 benchmark tasks by multiple metrics. Results show that FITNESS can outperform the state-of-the-art methods on bias mitigation while preserve the model's performance: it improved the model's fairness under all the scenarios while decreased the model's performance under only 26.67% of the scenarios. Additionally, FITNESS surpasses the Fairea Baseline in 96.72% cases, outperforming all methods we compared.
CATNet: Cross-event Attention-based Time-aware Network for Medical Event Prediction
Apr 29, 2022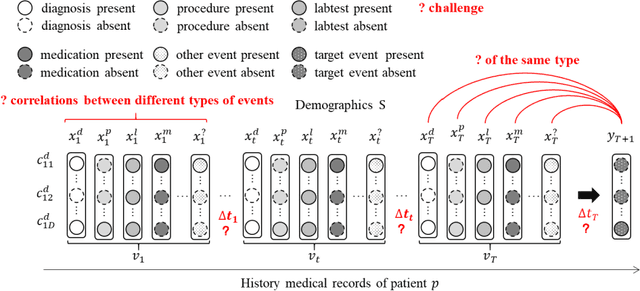

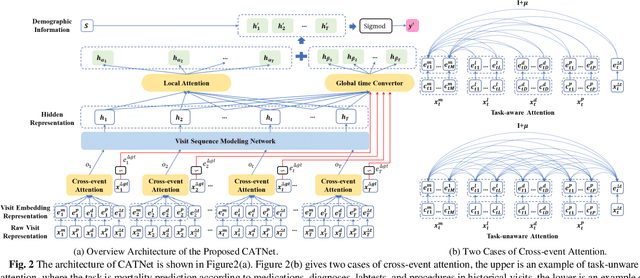
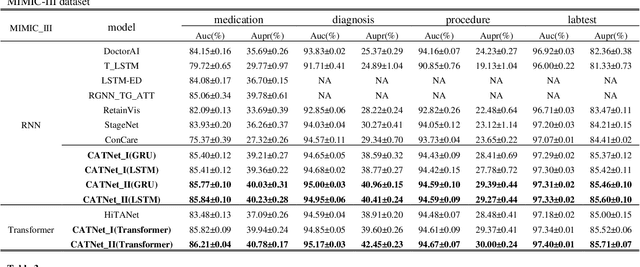
Abstract:Medical event prediction (MEP) is a fundamental task in the medical domain, which needs to predict medical events, including medications, diagnosis codes, laboratory tests, procedures, outcomes, and so on, according to historical medical records. The task is challenging as medical data is a type of complex time series data with heterogeneous and temporal irregular characteristics. Many machine learning methods that consider the two characteristics have been proposed for medical event prediction. However, most of them consider the two characteristics separately and ignore the correlations among different types of medical events, especially relations between historical medical events and target medical events. In this paper, we propose a novel neural network based on attention mechanism, called cross-event attention-based time-aware network (CATNet), for medical event prediction. It is a time-aware, event-aware and task-adaptive method with the following advantages: 1) modeling heterogeneous information and temporal information in a unified way and considering temporal irregular characteristics locally and globally respectively, 2) taking full advantage of correlations among different types of events via cross-event attention. Experiments on two public datasets (MIMIC-III and eICU) show CATNet can be adaptive with different MEP tasks and outperforms other state-of-the-art methods on various MEP tasks. The source code of CATNet will be released after this manuscript is accepted.
Two heads are better than one: Enhancing medical representations by pre-training over structured and unstructured electronic health records
Jan 25, 2022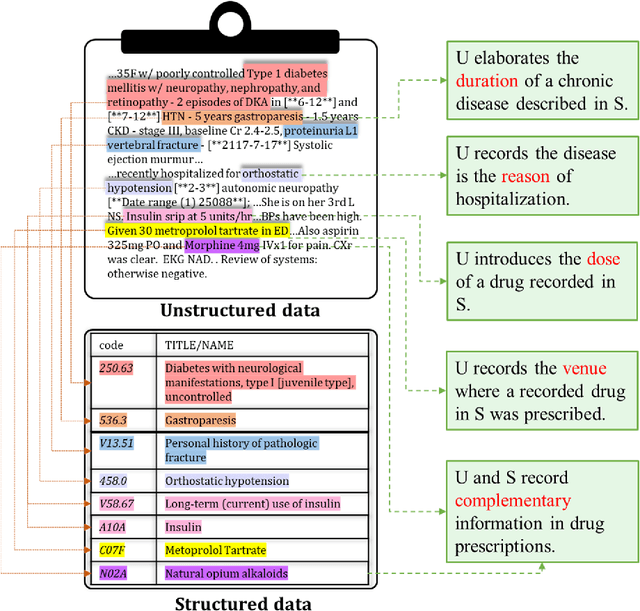
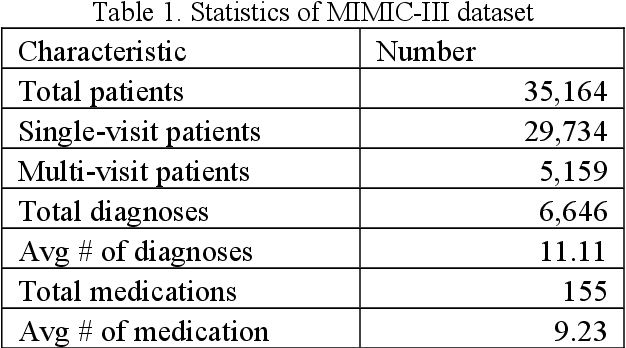
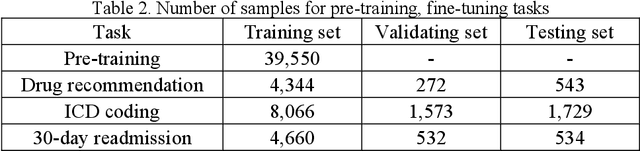
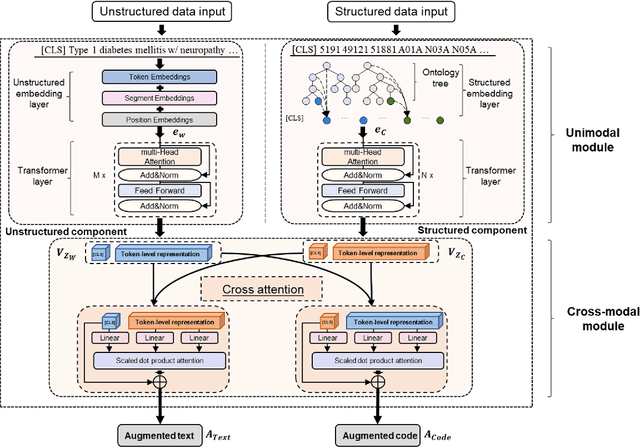
Abstract:The massive context of electronic health records (EHRs) has created enormous potentials for improving healthcare, among which structured (coded) data and unstructured (text) data are two important textual modalities. They do not exist in isolation and can complement each other in most real-life clinical scenarios. Most existing researches in medical informatics, however, either only focus on a particular modality or straightforwardly concatenate the information from different modalities, which ignore the interaction and information sharing between them. To address these issues, we proposed a unified deep learning-based medical pre-trained language model, named UMM-PLM, to automatically learn representative features from multimodal EHRs that consist of both structured data and unstructured data. Specifically, we first developed parallel unimodal information representation modules to capture the unimodal-specific characteristic, where unimodal representations were learned from each data source separately. A cross-modal module was further introduced to model the interactions between different modalities. We pre-trained the model on a large EHRs dataset containing both structured data and unstructured data and verified the effectiveness of the model on three downstream clinical tasks, i.e., medication recommendation, 30-day readmission and ICD coding through extensive experiments. The results demonstrate the power of UMM-PLM compared with benchmark methods and state-of-the-art baselines. Analyses show that UMM-PLM can effectively concern with multimodal textual information and has the potential to provide more comprehensive interpretations for clinical decision making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge