Yixi Xu
Influence of High-Performance Image-to-Image Translation Networks on Clinical Visual Assessment and Outcome Prediction: Utilizing Ultrasound to MRI Translation in Prostate Cancer
Jan 30, 2025



Abstract:Purpose: This study examines the core traits of image-to-image translation (I2I) networks, focusing on their effectiveness and adaptability in everyday clinical settings. Methods: We have analyzed data from 794 patients diagnosed with prostate cancer (PCa), using ten prominent 2D/3D I2I networks to convert ultrasound (US) images into MRI scans. We also introduced a new analysis of Radiomic features (RF) via the Spearman correlation coefficient to explore whether networks with high performance (SSIM>85%) could detect subtle RFs. Our study further examined synthetic images by 7 invited physicians. As a final evaluation study, we have investigated the improvement that are achieved using the synthetic MRI data on two traditional machine learning and one deep learning method. Results: In quantitative assessment, 2D-Pix2Pix network substantially outperformed the other 7 networks, with an average SSIM~0.855. The RF analysis revealed that 76 out of 186 RFs were identified using the 2D-Pix2Pix algorithm alone, although half of the RFs were lost during the translation process. A detailed qualitative review by 7 medical doctors noted a deficiency in low-level feature recognition in I2I tasks. Furthermore, the study found that synthesized image-based classification outperformed US image-based classification with an average accuracy and AUC~0.93. Conclusion: This study showed that while 2D-Pix2Pix outperformed cutting-edge networks in low-level feature discovery and overall error and similarity metrics, it still requires improvement in low-level feature performance, as highlighted by Group 3. Further, the study found using synthetic image-based classification outperformed original US image-based methods.
Biological and Radiological Dictionary of Radiomics Features: Addressing Understandable AI Issues in Personalized Prostate Cancer; Dictionary version PM1.0
Dec 14, 2024



Abstract:This study investigates the connection between visual semantic features in PI-RADS and associated risk factors, moving beyond abnormal imaging findings by creating a standardized dictionary of biological/radiological radiomics features (RFs). Using multiparametric prostate MRI sequences (T2-weighted imaging [T2WI], diffusion-weighted imaging [DWI], and apparent diffusion coefficient [ADC]), six interpretable and seven complex classifiers, combined with nine feature selection algorithms (FSAs), were applied to segmented lesions to predict UCLA scores. Combining T2WI, DWI, and ADC with FSAs such as ANOVA F-test, Correlation Coefficient, and Fisher Score, and utilizing logistic regression, identified key features: the 90th percentile from T2WI (hypo-intensity linked to cancer risk), variance from T2WI (lesion heterogeneity), shape metrics like Least Axis Length and Surface Area to Volume ratio from ADC (lesion compactness), and Run Entropy from ADC (texture consistency). This approach achieved an average accuracy of 0.78, outperforming single-sequence methods (p < 0.05). The developed dictionary provides a common language, fostering collaboration between clinical professionals and AI developers to enable trustworthy, interpretable AI for reliable clinical decisions.
IgCONDA-PET: Implicitly-Guided Counterfactual Diffusion for Detecting Anomalies in PET Images
Apr 30, 2024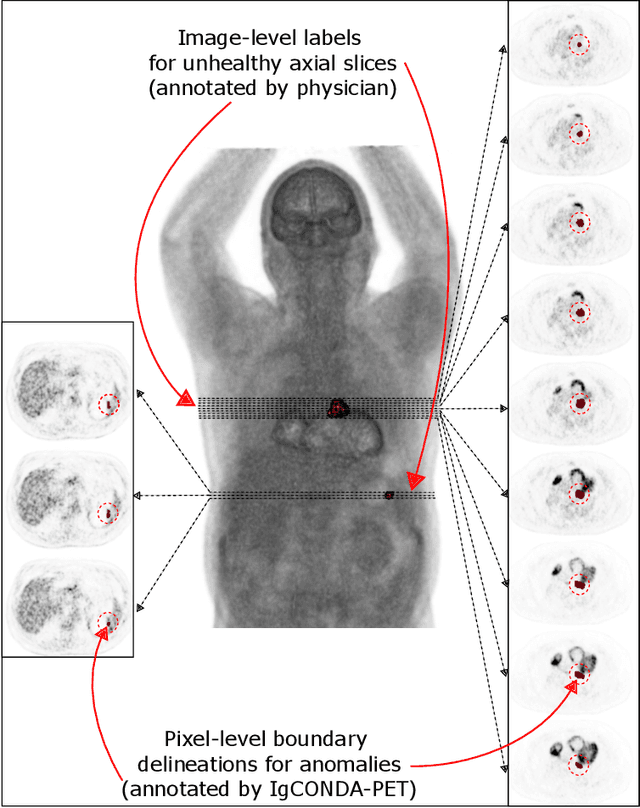
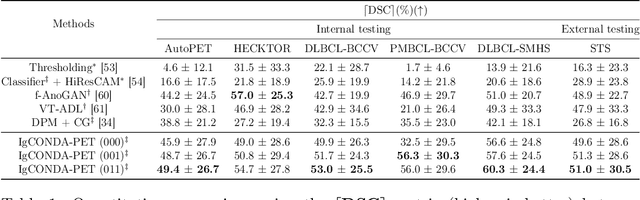
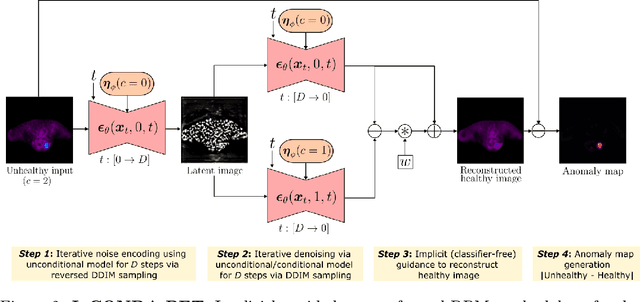
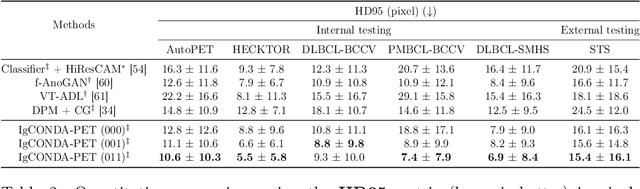
Abstract:Minimizing the need for pixel-level annotated data for training PET anomaly segmentation networks is crucial, particularly due to time and cost constraints related to expert annotations. Current un-/weakly-supervised anomaly detection methods rely on autoencoder or generative adversarial networks trained only on healthy data, although these are more challenging to train. In this work, we present a weakly supervised and Implicitly guided COuNterfactual diffusion model for Detecting Anomalies in PET images, branded as IgCONDA-PET. The training is conditioned on image class labels (healthy vs. unhealthy) along with implicit guidance to generate counterfactuals for an unhealthy image with anomalies. The counterfactual generation process synthesizes the healthy counterpart for a given unhealthy image, and the difference between the two facilitates the identification of anomaly locations. The code is available at: https://github.com/igcondapet/IgCONDA-PET.git
Do High-Performance Image-to-Image Translation Networks Enable the Discovery of Radiomic Features? Application to MRI Synthesis from Ultrasound in Prostate Cancer
Mar 27, 2024Abstract:This study investigates the foundational characteristics of image-to-image translation networks, specifically examining their suitability and transferability within the context of routine clinical environments, despite achieving high levels of performance, as indicated by a Structural Similarity Index (SSIM) exceeding 0.95. The evaluation study was conducted using data from 794 patients diagnosed with Prostate cancer. To synthesize MRI from Ultrasound images, we employed five widely recognized image to image translation networks in medical imaging: 2DPix2Pix, 2DCycleGAN, 3DCycleGAN, 3DUNET, and 3DAutoEncoder. For quantitative assessment, we report four prevalent evaluation metrics Mean Absolute Error, Mean Square Error, Structural Similarity Index (SSIM), and Peak Signal to Noise Ratio. Moreover, a complementary analysis employing Radiomic features (RF) via Spearman correlation coefficient was conducted to investigate, for the first time, whether networks achieving high performance, SSIM greater than 0.9, could identify low-level RFs. The RF analysis showed 76 features out of 186 RFs were discovered via just 2DPix2Pix algorithm while half of RFs were lost in the translation process. Finally, a detailed qualitative assessment by five medical doctors indicated a lack of low level feature discovery in image to image translation tasks.
A slice classification neural network for automated classification of axial PET/CT slices from a multi-centric lymphoma dataset
Mar 11, 2024Abstract:Automated slice classification is clinically relevant since it can be incorporated into medical image segmentation workflows as a preprocessing step that would flag slices with a higher probability of containing tumors, thereby directing physicians attention to the important slices. In this work, we train a ResNet-18 network to classify axial slices of lymphoma PET/CT images (collected from two institutions) depending on whether the slice intercepted a tumor (positive slice) in the 3D image or if the slice did not (negative slice). Various instances of the network were trained on 2D axial datasets created in different ways: (i) slice-level split and (ii) patient-level split; inputs of different types were used: (i) only PET slices and (ii) concatenated PET and CT slices; and different training strategies were employed: (i) center-aware (CAW) and (ii) center-agnostic (CAG). Model performances were compared using the area under the receiver operating characteristic curve (AUROC) and the area under the precision-recall curve (AUPRC), and various binary classification metrics. We observe and describe a performance overestimation in the case of slice-level split as compared to the patient-level split training. The model trained using patient-level split data with the network input containing only PET slices in the CAG training regime was the best performing/generalizing model on a majority of metrics. Our models were additionally more closely compared using the sensitivity metric on the positive slices from their respective test sets.
* 10 pages, 6 figures, 2 tables
Comprehensive Evaluation and Insights into the Use of Deep Neural Networks to Detect and Quantify Lymphoma Lesions in PET/CT Images
Nov 16, 2023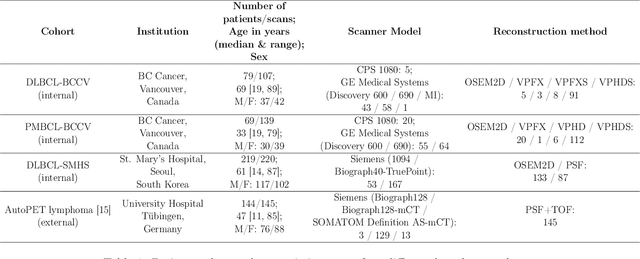
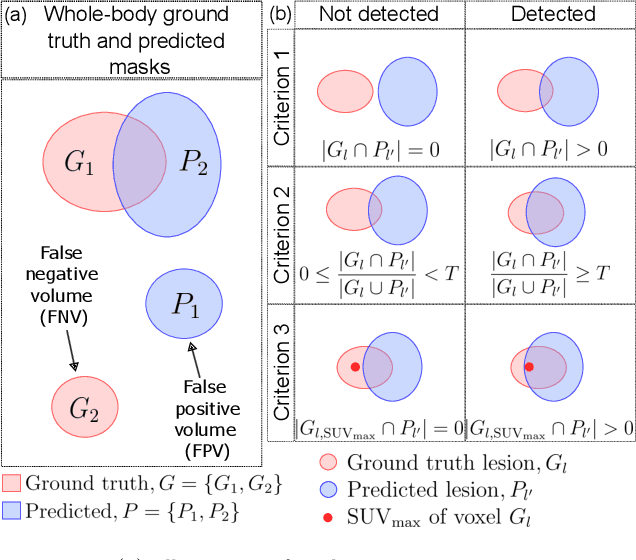
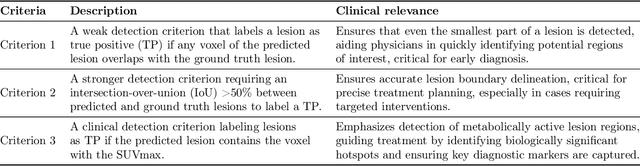
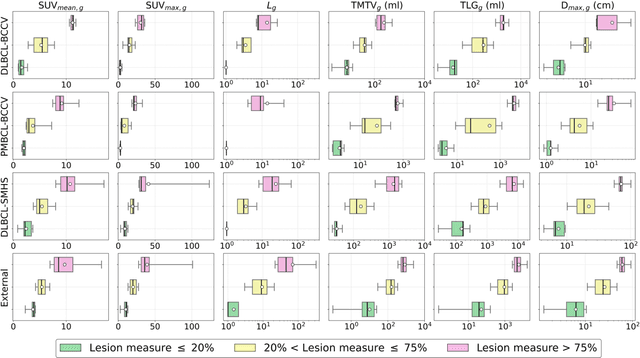
Abstract:This study performs comprehensive evaluation of four neural network architectures (UNet, SegResNet, DynUNet, and SwinUNETR) for lymphoma lesion segmentation from PET/CT images. These networks were trained, validated, and tested on a diverse, multi-institutional dataset of 611 cases. Internal testing (88 cases; total metabolic tumor volume (TMTV) range [0.52, 2300] ml) showed SegResNet as the top performer with a median Dice similarity coefficient (DSC) of 0.76 and median false positive volume (FPV) of 4.55 ml; all networks had a median false negative volume (FNV) of 0 ml. On the unseen external test set (145 cases with TMTV range: [0.10, 2480] ml), SegResNet achieved the best median DSC of 0.68 and FPV of 21.46 ml, while UNet had the best FNV of 0.41 ml. We assessed reproducibility of six lesion measures, calculated their prediction errors, and examined DSC performance in relation to these lesion measures, offering insights into segmentation accuracy and clinical relevance. Additionally, we introduced three lesion detection criteria, addressing the clinical need for identifying lesions, counting them, and segmenting based on metabolic characteristics. We also performed expert intra-observer variability analysis revealing the challenges in segmenting ``easy'' vs. ``hard'' cases, to assist in the development of more resilient segmentation algorithms. Finally, we performed inter-observer agreement assessment underscoring the importance of a standardized ground truth segmentation protocol involving multiple expert annotators. Code is available at: https://github.com/microsoft/lymphoma-segmentation-dnn
MACE: A Flexible Framework for Membership Privacy Estimation in Generative Models
Sep 11, 2020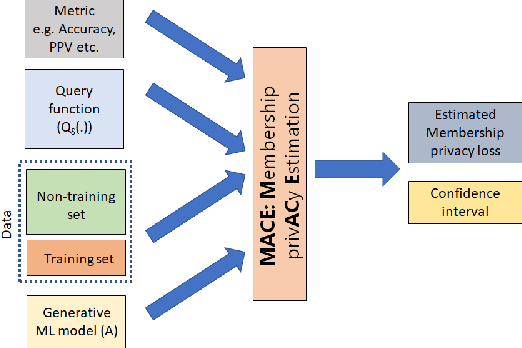
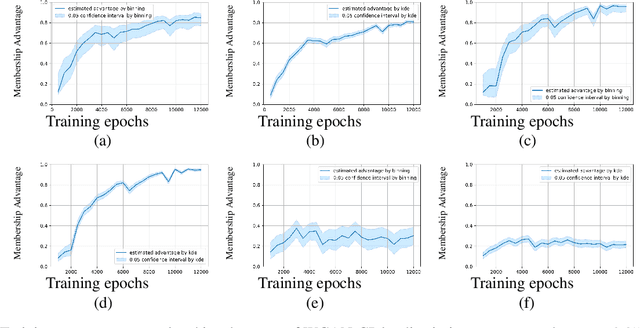
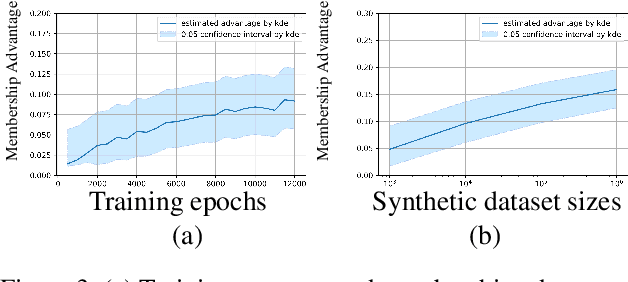
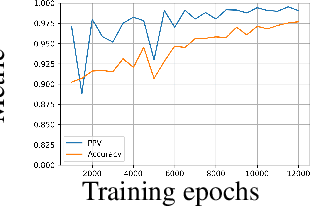
Abstract:Generative models are widely used for publishing synthetic datasets. Despite practical successes, recent works have shown some generative models may leak privacy of the data that have been used during training. Membership inference attacks aim to determine whether a sample has been used in the training set given query access to the model API. Despite recent work in this area, many of the attacks designed against generative models require very specific attributes from the learned models (e.g. discriminator scores, generated images, etc.). Furthermore, many of these attacks are heuristic and do not provide effective bounds for privacy loss. In this work, we formally study the membership privacy leakage risk of generative models. Specifically, we formulate membership privacy as a statistical divergence between training samples and hold-out samples, and propose sample-based methods to estimate this divergence. Unlike previous works, our proposed metric and estimators make realistic and flexible assumptions. First, we use a generalizable metric as an alternative to accuracy, since practical model training often leads to imbalanced train/hold-out splits. Second, our estimators are capable of estimating statistical divergence using any scalar or vector valued attributes from the learned model instead of very specific attributes. Furthermore, we show a connection to differential privacy. This allows our proposed estimators to provide a data-driven certificate to understand the privacy budget needed for differentially private generative models. We demonstrate the utility of our framework through experimental demonstrations on different generative models using various model attributes yielding some new insights about membership leakage and vulnerabilities of models.
Protecting GANs against privacy attacks by preventing overfitting
Jan 03, 2020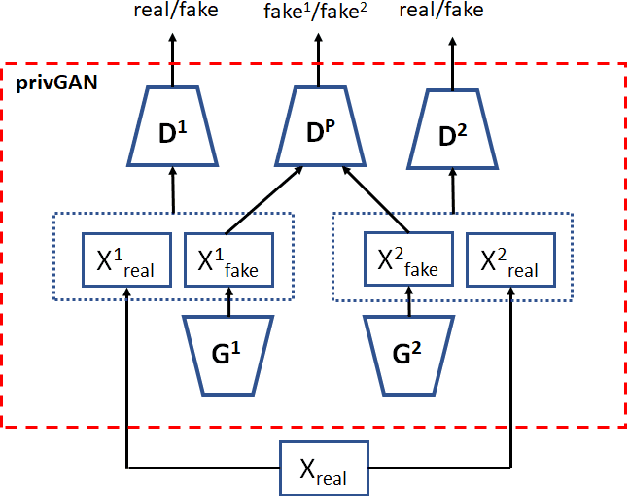

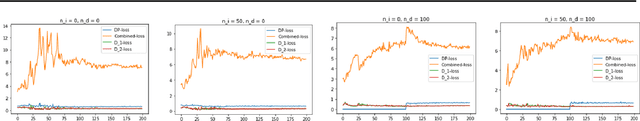
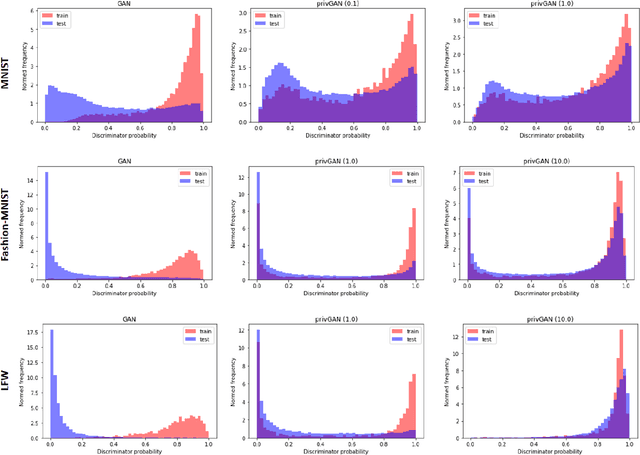
Abstract:Generative Adversarial Networks (GANs) have made releasing of synthetic images a viable approach to share data without releasing the original dataset. It has been shown that such synthetic data can be used for a variety of downstream tasks such as training classifiers that would otherwise require the original dataset to be shared. However, recent work has shown that the GAN models and their synthetically generated data can be used to infer the training set membership by an adversary who has access to the entire dataset and some auxiliary information. Here we develop a new GAN architecture (privGAN) which provides protection against this mode of attack while leading to negligible loss in downstream performances. Our architecture explicitly prevents overfitting to the training set thereby providing implicit protection against white-box attacks. The main contributions of this paper are: i) we propose a novel GAN architecture that can generate synthetic data in a privacy preserving manner and demonstrate the effectiveness of our model against white--box attacks on several benchmark datasets, ii) we provide a theoretical understanding of the optimal solution of the GAN loss function, iii) we demonstrate on two common benchmark datasets that synthetic images generated by privGAN lead to negligible loss in downstream performance when compared against non--private GANs. While we have focosued on benchmarking privGAN exclusively of image datasets, the architecture of privGAN is not exclusive to image datasets and can be easily extended to other types of datasets.
Understanding Weight Normalized Deep Neural Networks with Rectified Linear Units
Oct 15, 2018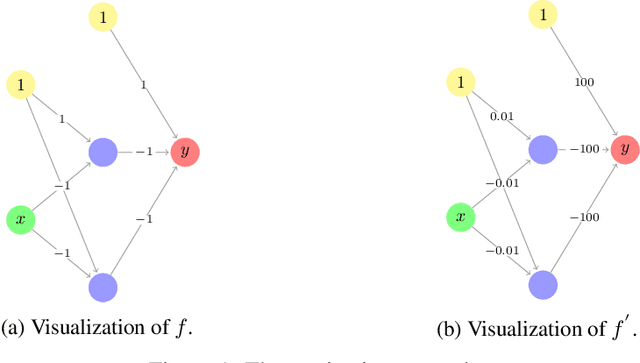
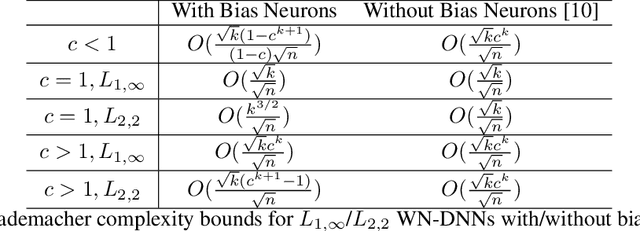
Abstract:This paper presents a general framework for norm-based capacity control for $L_{p,q}$ weight normalized deep neural networks. We establish the upper bound on the Rademacher complexities of this family. With an $L_{p,q}$ normalization where $q\le p^*$, and $1/p+1/p^{*}=1$, we discuss properties of a width-independent capacity control, which only depends on depth by a square root term. We further analyze the approximation properties of $L_{p,q}$ weight normalized deep neural networks. In particular, for an $L_{1,\infty}$ weight normalized network, the approximation error can be controlled by the $L_1$ norm of the output layer, and the corresponding generalization error only depends on the architecture by the square root of the depth.
On the Statistical Efficiency of Compositional Nonparametric Prediction
Oct 20, 2017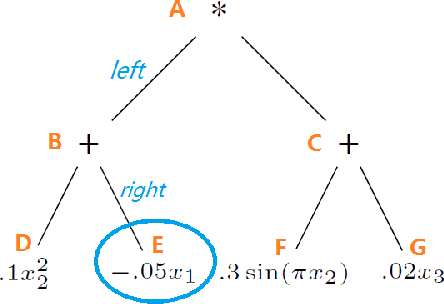
Abstract:In this paper, we propose a compositional nonparametric method in which a model is expressed as a labeled binary tree of $2k+1$ nodes, where each node is either a summation, a multiplication, or the application of one of the $q$ basis functions to one of the $p$ covariates. We show that in order to recover a labeled binary tree from a given dataset, the sufficient number of samples is $O(k\log(pq)+\log(k!))$, and the necessary number of samples is $\Omega(k\log (pq)-\log(k!))$. We further propose a greedy algorithm for regression in order to validate our theoretical findings through synthetic experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge