Ingrid Bloise
A cascaded deep network for automated tumor detection and segmentation in clinical PET imaging of diffuse large B-cell lymphoma
Mar 11, 2024Abstract:Accurate detection and segmentation of diffuse large B-cell lymphoma (DLBCL) from PET images has important implications for estimation of total metabolic tumor volume, radiomics analysis, surgical intervention and radiotherapy. Manual segmentation of tumors in whole-body PET images is time-consuming, labor-intensive and operator-dependent. In this work, we develop and validate a fast and efficient three-step cascaded deep learning model for automated detection and segmentation of DLBCL tumors from PET images. As compared to a single end-to-end network for segmentation of tumors in whole-body PET images, our three-step model is more effective (improves 3D Dice score from 58.9% to 78.1%) since each of its specialized modules, namely the slice classifier, the tumor detector and the tumor segmentor, can be trained independently to a high degree of skill to carry out a specific task, rather than a single network with suboptimal performance on overall segmentation.
* 8 pages, 3 figures, 3 tables
A slice classification neural network for automated classification of axial PET/CT slices from a multi-centric lymphoma dataset
Mar 11, 2024Abstract:Automated slice classification is clinically relevant since it can be incorporated into medical image segmentation workflows as a preprocessing step that would flag slices with a higher probability of containing tumors, thereby directing physicians attention to the important slices. In this work, we train a ResNet-18 network to classify axial slices of lymphoma PET/CT images (collected from two institutions) depending on whether the slice intercepted a tumor (positive slice) in the 3D image or if the slice did not (negative slice). Various instances of the network were trained on 2D axial datasets created in different ways: (i) slice-level split and (ii) patient-level split; inputs of different types were used: (i) only PET slices and (ii) concatenated PET and CT slices; and different training strategies were employed: (i) center-aware (CAW) and (ii) center-agnostic (CAG). Model performances were compared using the area under the receiver operating characteristic curve (AUROC) and the area under the precision-recall curve (AUPRC), and various binary classification metrics. We observe and describe a performance overestimation in the case of slice-level split as compared to the patient-level split training. The model trained using patient-level split data with the network input containing only PET slices in the CAG training regime was the best performing/generalizing model on a majority of metrics. Our models were additionally more closely compared using the sensitivity metric on the positive slices from their respective test sets.
* 10 pages, 6 figures, 2 tables
Observer study-based evaluation of TGAN architecture used to generate oncological PET images
Nov 28, 2023Abstract:The application of computer-vision algorithms in medical imaging has increased rapidly in recent years. However, algorithm training is challenging due to limited sample sizes, lack of labeled samples, as well as privacy concerns regarding data sharing. To address these issues, we previously developed (Bergen et al. 2022) a synthetic PET dataset for Head and Neck (H and N) cancer using the temporal generative adversarial network (TGAN) architecture and evaluated its performance segmenting lesions and identifying radiomics features in synthesized images. In this work, a two-alternative forced-choice (2AFC) observer study was performed to quantitatively evaluate the ability of human observers to distinguish between real and synthesized oncological PET images. In the study eight trained readers, including two board-certified nuclear medicine physicians, read 170 real/synthetic image pairs presented as 2D-transaxial using a dedicated web app. For each image pair, the observer was asked to identify the real image and input their confidence level with a 5-point Likert scale. P-values were computed using the binomial test and Wilcoxon signed-rank test. A heat map was used to compare the response accuracy distribution for the signed-rank test. Response accuracy for all observers ranged from 36.2% [27.9-44.4] to 63.1% [54.8-71.3]. Six out of eight observers did not identify the real image with statistical significance, indicating that the synthetic dataset was reasonably representative of oncological PET images. Overall, this study adds validity to the realism of our simulated H&N cancer dataset, which may be implemented in the future to train AI algorithms while favoring patient confidentiality and privacy protection.
Comprehensive Evaluation and Insights into the Use of Deep Neural Networks to Detect and Quantify Lymphoma Lesions in PET/CT Images
Nov 16, 2023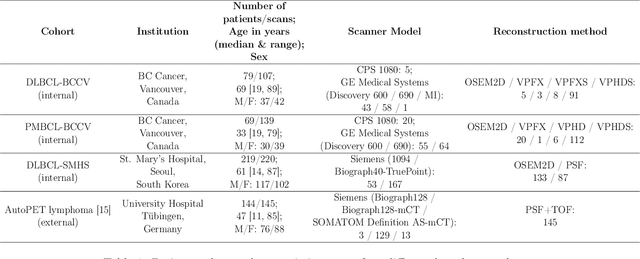
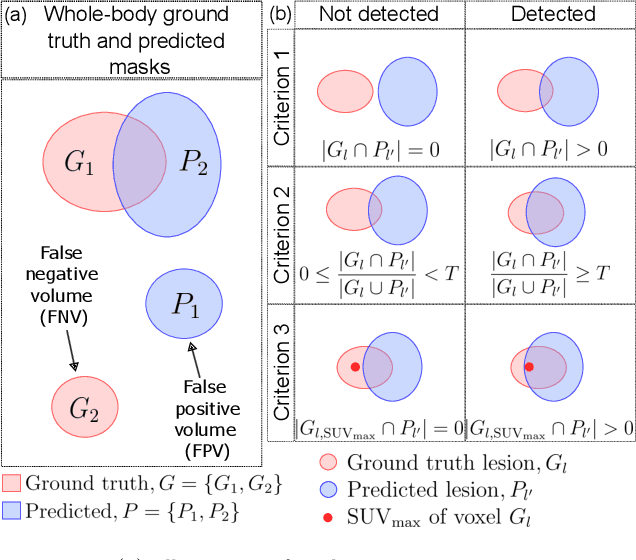
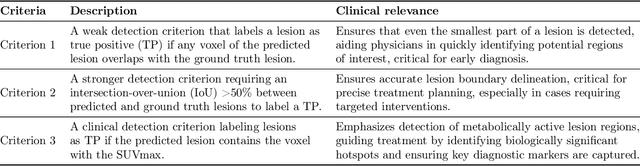
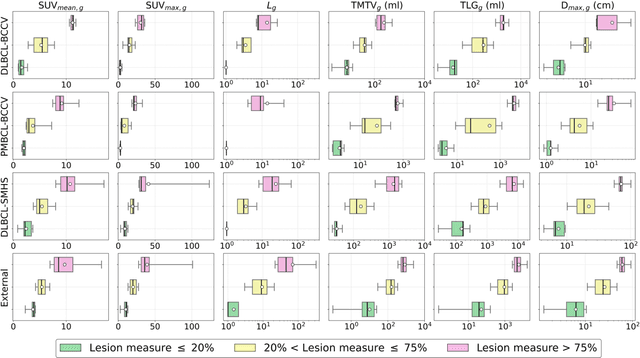
Abstract:This study performs comprehensive evaluation of four neural network architectures (UNet, SegResNet, DynUNet, and SwinUNETR) for lymphoma lesion segmentation from PET/CT images. These networks were trained, validated, and tested on a diverse, multi-institutional dataset of 611 cases. Internal testing (88 cases; total metabolic tumor volume (TMTV) range [0.52, 2300] ml) showed SegResNet as the top performer with a median Dice similarity coefficient (DSC) of 0.76 and median false positive volume (FPV) of 4.55 ml; all networks had a median false negative volume (FNV) of 0 ml. On the unseen external test set (145 cases with TMTV range: [0.10, 2480] ml), SegResNet achieved the best median DSC of 0.68 and FPV of 21.46 ml, while UNet had the best FNV of 0.41 ml. We assessed reproducibility of six lesion measures, calculated their prediction errors, and examined DSC performance in relation to these lesion measures, offering insights into segmentation accuracy and clinical relevance. Additionally, we introduced three lesion detection criteria, addressing the clinical need for identifying lesions, counting them, and segmenting based on metabolic characteristics. We also performed expert intra-observer variability analysis revealing the challenges in segmenting ``easy'' vs. ``hard'' cases, to assist in the development of more resilient segmentation algorithms. Finally, we performed inter-observer agreement assessment underscoring the importance of a standardized ground truth segmentation protocol involving multiple expert annotators. Code is available at: https://github.com/microsoft/lymphoma-segmentation-dnn
Semi-supervised learning towards automated segmentation of PET images with limited annotations: Application to lymphoma patients
Dec 24, 2022Abstract:The time-consuming task of manual segmentation challenges routine systematic quantification of disease burden. Convolutional neural networks (CNNs) hold significant promise to reliably identify locations and boundaries of tumors from PET scans. We aimed to leverage the need for annotated data via semi-supervised approaches, with application to PET images of diffuse large B-cell lymphoma (DLBCL) and primary mediastinal large B-cell lymphoma (PMBCL). We analyzed 18F-FDG PET images of 292 patients with PMBCL (n=104) and DLBCL (n=188) (n=232 for training and validation, and n=60 for external testing). We employed FCM and MS losses for training a 3D U-Net with different levels of supervision: i) fully supervised methods with labeled FCM (LFCM) as well as Unified focal and Dice loss functions, ii) unsupervised methods with Robust FCM (RFCM) and Mumford-Shah (MS) loss functions, and iii) Semi-supervised methods based on FCM (RFCM+LFCM), as well as MS loss in combination with supervised Dice loss (MS+Dice). Unified loss function yielded higher Dice score (mean +/- standard deviation (SD)) (0.73 +/- 0.03; 95% CI, 0.67-0.8) compared to Dice loss (p-value<0.01). Semi-supervised (RFCM+alpha*LFCM) with alpha=0.3 showed the best performance, with a Dice score of 0.69 +/- 0.03 (95% CI, 0.45-0.77) outperforming (MS+alpha*Dice) for any supervision level (any alpha) (p<0.01). The best performer among (MS+alpha*Dice) semi-supervised approaches with alpha=0.2 showed a Dice score of 0.60 +/- 0.08 (95% CI, 0.44-0.76) compared to another supervision level in this semi-supervised approach (p<0.01). Semi-supervised learning via FCM loss (RFCM+alpha*LFCM) showed improved performance compared to supervised approaches. Considering the time-consuming nature of expert manual delineations and intra-observer variabilities, semi-supervised approaches have significant potential for automated segmentation workflows.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge