Natalia Dubljevic
A cascaded deep network for automated tumor detection and segmentation in clinical PET imaging of diffuse large B-cell lymphoma
Mar 11, 2024Abstract:Accurate detection and segmentation of diffuse large B-cell lymphoma (DLBCL) from PET images has important implications for estimation of total metabolic tumor volume, radiomics analysis, surgical intervention and radiotherapy. Manual segmentation of tumors in whole-body PET images is time-consuming, labor-intensive and operator-dependent. In this work, we develop and validate a fast and efficient three-step cascaded deep learning model for automated detection and segmentation of DLBCL tumors from PET images. As compared to a single end-to-end network for segmentation of tumors in whole-body PET images, our three-step model is more effective (improves 3D Dice score from 58.9% to 78.1%) since each of its specialized modules, namely the slice classifier, the tumor detector and the tumor segmentor, can be trained independently to a high degree of skill to carry out a specific task, rather than a single network with suboptimal performance on overall segmentation.
* 8 pages, 3 figures, 3 tables
Tensor Radiomics: Paradigm for Systematic Incorporation of Multi-Flavoured Radiomics Features
Mar 12, 2022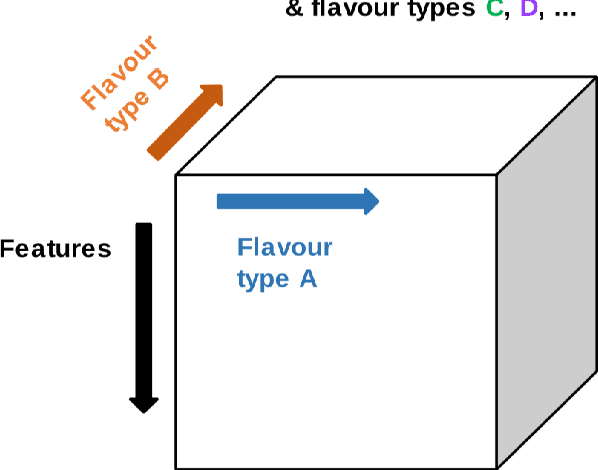
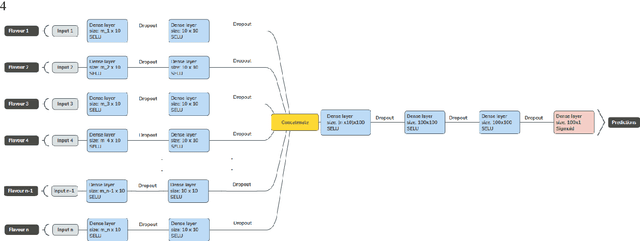
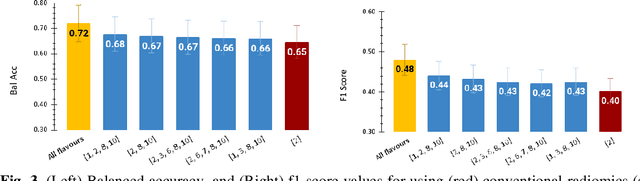
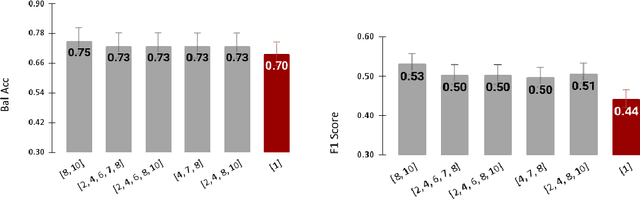
Abstract:Radiomics features extract quantitative information from medical images, towards the derivation of biomarkers for clinical tasks, such as diagnosis, prognosis, or treatment response assessment. Different image discretization parameters (e.g. bin number or size), convolutional filters, segmentation perturbation, or multi-modality fusion levels can be used to generate radiomics features and ultimately signatures. Commonly, only one set of parameters is used; resulting in only one value or flavour for a given RF. We propose tensor radiomics (TR) where tensors of features calculated with multiple combinations of parameters (i.e. flavours) are utilized to optimize the construction of radiomics signatures. We present examples of TR as applied to PET/CT, MRI, and CT imaging invoking machine learning or deep learning solutions, and reproducibility analyses: (1) TR via varying bin sizes on CT images of lung cancer and PET-CT images of head & neck cancer (HNC) for overall survival prediction. A hybrid deep neural network, referred to as TR-Net, along with two ML-based flavour fusion methods showed improved accuracy compared to regular rediomics features. (2) TR built from different segmentation perturbations and different bin sizes for classification of late-stage lung cancer response to first-line immunotherapy using CT images. TR improved predicted patient responses. (3) TR via multi-flavour generated radiomics features in MR imaging showed improved reproducibility when compared to many single-flavour features. (4) TR via multiple PET/CT fusions in HNC. Flavours were built from different fusions using methods, such as Laplacian pyramids and wavelet transforms. TR improved overall survival prediction. Our results suggest that the proposed TR paradigm has the potential to improve performance capabilities in different medical imaging tasks.
Segmentation and Risk Score Prediction of Head and Neck Cancers in PET/CT Volumes with 3D U-Net and Cox Proportional Hazard Neural Networks
Feb 16, 2022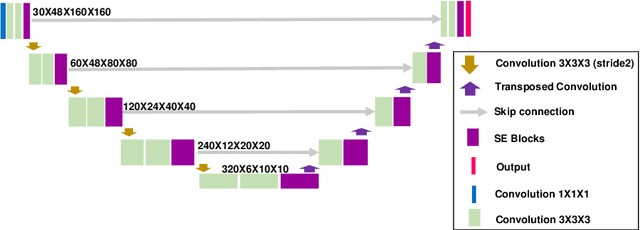
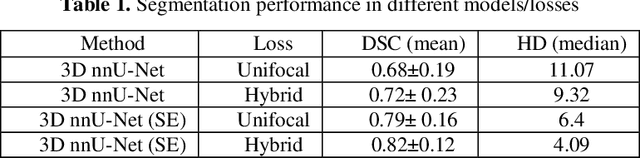
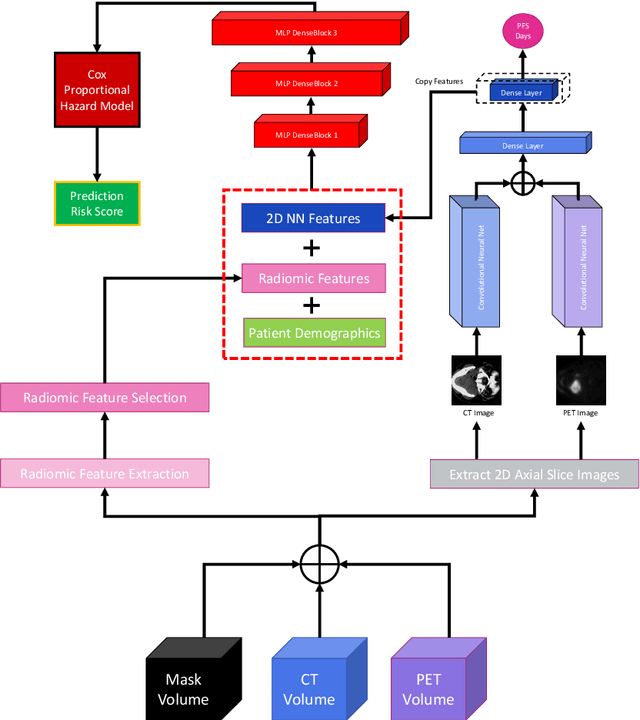
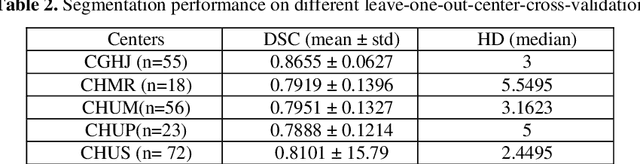
Abstract:We utilized a 3D nnU-Net model with residual layers supplemented by squeeze and excitation (SE) normalization for tumor segmentation from PET/CT images provided by the Head and Neck Tumor segmentation chal-lenge (HECKTOR). Our proposed loss function incorporates the Unified Fo-cal and Mumford-Shah losses to take the advantage of distribution, region, and boundary-based loss functions. The results of leave-one-out-center-cross-validation performed on different centers showed a segmentation performance of 0.82 average Dice score (DSC) and 3.16 median Hausdorff Distance (HD), and our results on the test set achieved 0.77 DSC and 3.01 HD. Following lesion segmentation, we proposed training a case-control proportional hazard Cox model with an MLP neural net backbone to predict the hazard risk score for each discrete lesion. This hazard risk prediction model (CoxCC) was to be trained on a number of PET/CT radiomic features extracted from the segmented lesions, patient and lesion demographics, and encoder features provided from the penultimate layer of a multi-input 2D PET/CT convolutional neural network tasked with predicting time-to-event for each lesion. A 10-fold cross-validated CoxCC model resulted in a c-index validation score of 0.89, and a c-index score of 0.61 on the HECKTOR challenge test dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge