Habib Zaidi
Unveiling and Bridging the Functional Perception Gap in MLLMs: Atomic Visual Alignment and Hierarchical Evaluation via PET-Bench
Jan 06, 2026Abstract:While Multimodal Large Language Models (MLLMs) have demonstrated remarkable proficiency in tasks such as abnormality detection and report generation for anatomical modalities, their capability in functional imaging remains largely unexplored. In this work, we identify and quantify a fundamental functional perception gap: the inability of current vision encoders to decode functional tracer biodistribution independent of morphological priors. Identifying Positron Emission Tomography (PET) as the quintessential modality to investigate this disconnect, we introduce PET-Bench, the first large-scale functional imaging benchmark comprising 52,308 hierarchical QA pairs from 9,732 multi-site, multi-tracer PET studies. Extensive evaluation of 19 state-of-the-art MLLMs reveals a critical safety hazard termed the Chain-of-Thought (CoT) hallucination trap. We observe that standard CoT prompting, widely considered to enhance reasoning, paradoxically decouples linguistic generation from visual evidence in PET, producing clinically fluent but factually ungrounded diagnoses. To resolve this, we propose Atomic Visual Alignment (AVA), a simple fine-tuning strategy that enforces the mastery of low-level functional perception prior to high-level diagnostic reasoning. Our results demonstrate that AVA effectively bridges the perception gap, transforming CoT from a source of hallucination into a robust inference tool and improving diagnostic accuracy by up to 14.83%. Code and data are available at https://github.com/yezanting/PET-Bench.
D-PerceptCT: Deep Perceptual Enhancement for Low-Dose CT Images
Nov 18, 2025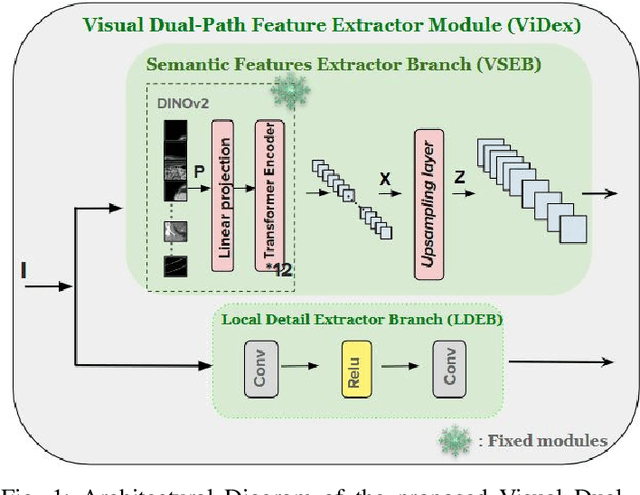
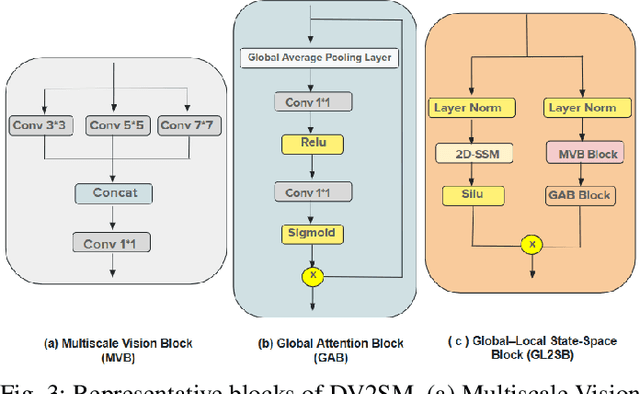
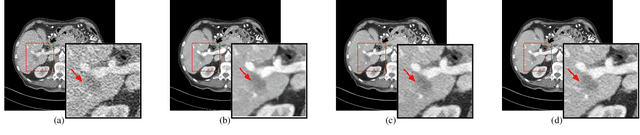
Abstract:Low Dose Computed Tomography (LDCT) is widely used as an imaging solution to aid diagnosis and other clinical tasks. However, this comes at the price of a deterioration in image quality due to the low dose of radiation used to reduce the risk of secondary cancer development. While some efficient methods have been proposed to enhance LDCT quality, many overestimate noise and perform excessive smoothing, leading to a loss of critical details. In this paper, we introduce D-PerceptCT, a novel architecture inspired by key principles of the Human Visual System (HVS) to enhance LDCT images. The objective is to guide the model to enhance or preserve perceptually relevant features, thereby providing radiologists with CT images where critical anatomical structures and fine pathological details are perceptu- ally visible. D-PerceptCT consists of two main blocks: 1) a Visual Dual-path Extractor (ViDex), which integrates semantic priors from a pretrained DINOv2 model with local spatial features, allowing the network to incorporate semantic-awareness during enhancement; (2) a Global-Local State-Space block that captures long-range information and multiscale features to preserve the important structures and fine details for diagnosis. In addition, we propose a novel deep perceptual loss, designated as the Deep Perceptual Relevancy Loss Function (DPRLF), which is inspired by human contrast sensitivity, to further emphasize perceptually important features. Extensive experiments on the Mayo2016 dataset demonstrate the effectiveness of D-PerceptCT method for LDCT enhancement, showing better preservation of structural and textural information within LDCT images compared to SOTA methods.
Artificial intelligence for simplified patient-centered dosimetry in radiopharmaceutical therapies
Oct 14, 2025Abstract:KEY WORDS: Artificial Intelligence (AI), Theranostics, Dosimetry, Radiopharmaceutical Therapy (RPT), Patient-friendly dosimetry KEY POINTS - The rapid evolution of radiopharmaceutical therapy (RPT) highlights the growing need for personalized and patient-centered dosimetry. - Artificial Intelligence (AI) offers solutions to the key limitations in current dosimetry calculations. - The main advances on AI for simplified dosimetry toward patient-friendly RPT are reviewed. - Future directions on the role of AI in RPT dosimetry are discussed.
From Claims to Evidence: A Unified Framework and Critical Analysis of CNN vs. Transformer vs. Mamba in Medical Image Segmentation
Mar 03, 2025



Abstract:While numerous architectures for medical image segmentation have been proposed, achieving competitive performance with state-of-the-art models networks such as nnUNet, still leave room for further innovation. In this work, we introduce nnUZoo, an open source benchmarking framework built upon nnUNet, which incorporates various deep learning architectures, including CNNs, Transformers, and Mamba-based models. Using this framework, we provide a fair comparison to demystify performance claims across different medical image segmentation tasks. Additionally, in an effort to enrich the benchmarking, we explored five new architectures based on Mamba and Transformers, collectively named X2Net, and integrated them into nnUZoo for further evaluation. The proposed models combine the features of conventional U2Net, nnUNet, CNN, Transformer, and Mamba layers and architectures, called X2Net (UNETR2Net (UNETR), SwT2Net (SwinTransformer), SS2D2Net (SwinUMamba), Alt1DM2Net (LightUMamba), and MambaND2Net (MambaND)). We extensively evaluate the performance of different models on six diverse medical image segmentation datasets, including microscopy, ultrasound, CT, MRI, and PET, covering various body parts, organs, and labels. We compare their performance, in terms of dice score and computational efficiency, against their baseline models, U2Net, and nnUNet. CNN models like nnUNet and U2Net demonstrated both speed and accuracy, making them effective choices for medical image segmentation tasks. Transformer-based models, while promising for certain imaging modalities, exhibited high computational costs. Proposed Mamba-based X2Net architecture (SS2D2Net) achieved competitive accuracy with no significantly difference from nnUNet and U2Net, while using fewer parameters. However, they required significantly longer training time, highlighting a trade-off between model efficiency and computational cost.
Thyroidiomics: An Automated Pipeline for Segmentation and Classification of Thyroid Pathologies from Scintigraphy Images
Jul 14, 2024



Abstract:The objective of this study was to develop an automated pipeline that enhances thyroid disease classification using thyroid scintigraphy images, aiming to decrease assessment time and increase diagnostic accuracy. Anterior thyroid scintigraphy images from 2,643 patients were collected and categorized into diffuse goiter (DG), multinodal goiter (MNG), and thyroiditis (TH) based on clinical reports, and then segmented by an expert. A ResUNet model was trained to perform auto-segmentation. Radiomic features were extracted from both physician (scenario 1) and ResUNet segmentations (scenario 2), followed by omitting highly correlated features using Spearman's correlation, and feature selection using Recursive Feature Elimination (RFE) with XGBoost as the core. All models were trained under leave-one-center-out cross-validation (LOCOCV) scheme, where nine instances of algorithms were iteratively trained and validated on data from eight centers and tested on the ninth for both scenarios separately. Segmentation performance was assessed using the Dice similarity coefficient (DSC), while classification performance was assessed using metrics, such as precision, recall, F1-score, accuracy, area under the Receiver Operating Characteristic (ROC AUC), and area under the precision-recall curve (PRC AUC). ResUNet achieved DSC values of 0.84$\pm$0.03, 0.71$\pm$0.06, and 0.86$\pm$0.02 for MNG, TH, and DG, respectively. Classification in scenario 1 achieved an accuracy of 0.76$\pm$0.04 and a ROC AUC of 0.92$\pm$0.02 while in scenario 2, classification yielded an accuracy of 0.74$\pm$0.05 and a ROC AUC of 0.90$\pm$0.02. The automated pipeline demonstrated comparable performance to physician segmentations on several classification metrics across different classes, effectively reducing assessment time while maintaining high diagnostic accuracy. Code available at: https://github.com/ahxmeds/thyroidiomics.git.
Segmentation-Free Outcome Prediction in Head and Neck Cancer: Deep Learning-based Feature Extraction from Multi-Angle Maximum Intensity Projections (MA-MIPs) of PET Images
May 02, 2024



Abstract:We introduce an innovative, simple, effective segmentation-free approach for outcome prediction in head \& neck cancer (HNC) patients. By harnessing deep learning-based feature extraction techniques and multi-angle maximum intensity projections (MA-MIPs) applied to Fluorodeoxyglucose Positron Emission Tomography (FDG-PET) volumes, our proposed method eliminates the need for manual segmentations of regions-of-interest (ROIs) such as primary tumors and involved lymph nodes. Instead, a state-of-the-art object detection model is trained to perform automatic cropping of the head and neck region on the PET volumes. A pre-trained deep convolutional neural network backbone is then utilized to extract deep features from MA-MIPs obtained from 72 multi-angel axial rotations of the cropped PET volumes. These deep features extracted from multiple projection views of the PET volumes are then aggregated and fused, and employed to perform recurrence-free survival analysis on a cohort of 489 HNC patients. The proposed approach outperforms the best performing method on the target dataset for the task of recurrence-free survival analysis. By circumventing the manual delineation of the malignancies on the FDG PET-CT images, our approach eliminates the dependency on subjective interpretations and highly enhances the reproducibility of the proposed survival analysis method.
Semi-supervised learning towards automated segmentation of PET images with limited annotations: Application to lymphoma patients
Dec 24, 2022Abstract:The time-consuming task of manual segmentation challenges routine systematic quantification of disease burden. Convolutional neural networks (CNNs) hold significant promise to reliably identify locations and boundaries of tumors from PET scans. We aimed to leverage the need for annotated data via semi-supervised approaches, with application to PET images of diffuse large B-cell lymphoma (DLBCL) and primary mediastinal large B-cell lymphoma (PMBCL). We analyzed 18F-FDG PET images of 292 patients with PMBCL (n=104) and DLBCL (n=188) (n=232 for training and validation, and n=60 for external testing). We employed FCM and MS losses for training a 3D U-Net with different levels of supervision: i) fully supervised methods with labeled FCM (LFCM) as well as Unified focal and Dice loss functions, ii) unsupervised methods with Robust FCM (RFCM) and Mumford-Shah (MS) loss functions, and iii) Semi-supervised methods based on FCM (RFCM+LFCM), as well as MS loss in combination with supervised Dice loss (MS+Dice). Unified loss function yielded higher Dice score (mean +/- standard deviation (SD)) (0.73 +/- 0.03; 95% CI, 0.67-0.8) compared to Dice loss (p-value<0.01). Semi-supervised (RFCM+alpha*LFCM) with alpha=0.3 showed the best performance, with a Dice score of 0.69 +/- 0.03 (95% CI, 0.45-0.77) outperforming (MS+alpha*Dice) for any supervision level (any alpha) (p<0.01). The best performer among (MS+alpha*Dice) semi-supervised approaches with alpha=0.2 showed a Dice score of 0.60 +/- 0.08 (95% CI, 0.44-0.76) compared to another supervision level in this semi-supervised approach (p<0.01). Semi-supervised learning via FCM loss (RFCM+alpha*LFCM) showed improved performance compared to supervised approaches. Considering the time-consuming nature of expert manual delineations and intra-observer variabilities, semi-supervised approaches have significant potential for automated segmentation workflows.
Tensor Radiomics: Paradigm for Systematic Incorporation of Multi-Flavoured Radiomics Features
Mar 12, 2022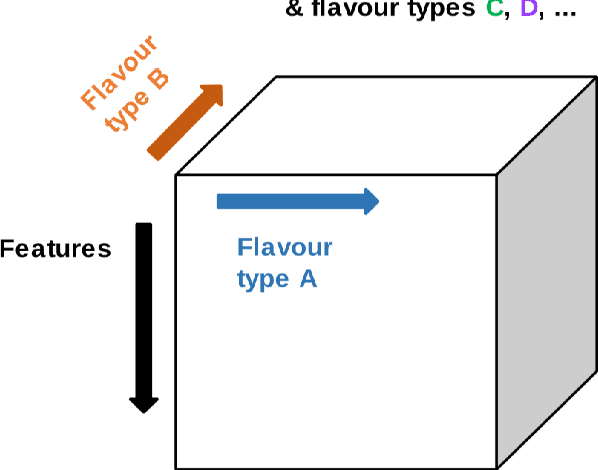
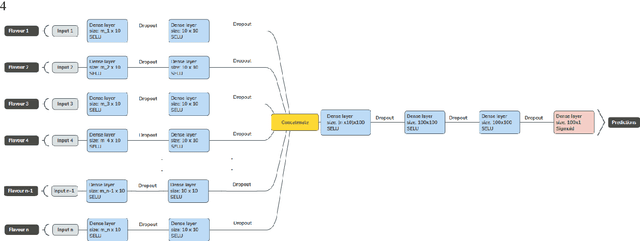
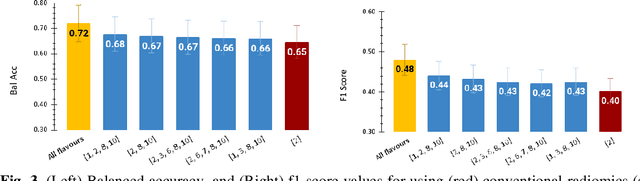
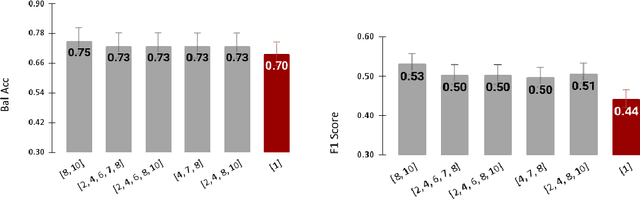
Abstract:Radiomics features extract quantitative information from medical images, towards the derivation of biomarkers for clinical tasks, such as diagnosis, prognosis, or treatment response assessment. Different image discretization parameters (e.g. bin number or size), convolutional filters, segmentation perturbation, or multi-modality fusion levels can be used to generate radiomics features and ultimately signatures. Commonly, only one set of parameters is used; resulting in only one value or flavour for a given RF. We propose tensor radiomics (TR) where tensors of features calculated with multiple combinations of parameters (i.e. flavours) are utilized to optimize the construction of radiomics signatures. We present examples of TR as applied to PET/CT, MRI, and CT imaging invoking machine learning or deep learning solutions, and reproducibility analyses: (1) TR via varying bin sizes on CT images of lung cancer and PET-CT images of head & neck cancer (HNC) for overall survival prediction. A hybrid deep neural network, referred to as TR-Net, along with two ML-based flavour fusion methods showed improved accuracy compared to regular rediomics features. (2) TR built from different segmentation perturbations and different bin sizes for classification of late-stage lung cancer response to first-line immunotherapy using CT images. TR improved predicted patient responses. (3) TR via multi-flavour generated radiomics features in MR imaging showed improved reproducibility when compared to many single-flavour features. (4) TR via multiple PET/CT fusions in HNC. Flavours were built from different fusions using methods, such as Laplacian pyramids and wavelet transforms. TR improved overall survival prediction. Our results suggest that the proposed TR paradigm has the potential to improve performance capabilities in different medical imaging tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge