Seyed Rasoul Zakavi
Thyroidiomics: An Automated Pipeline for Segmentation and Classification of Thyroid Pathologies from Scintigraphy Images
Jul 14, 2024



Abstract:The objective of this study was to develop an automated pipeline that enhances thyroid disease classification using thyroid scintigraphy images, aiming to decrease assessment time and increase diagnostic accuracy. Anterior thyroid scintigraphy images from 2,643 patients were collected and categorized into diffuse goiter (DG), multinodal goiter (MNG), and thyroiditis (TH) based on clinical reports, and then segmented by an expert. A ResUNet model was trained to perform auto-segmentation. Radiomic features were extracted from both physician (scenario 1) and ResUNet segmentations (scenario 2), followed by omitting highly correlated features using Spearman's correlation, and feature selection using Recursive Feature Elimination (RFE) with XGBoost as the core. All models were trained under leave-one-center-out cross-validation (LOCOCV) scheme, where nine instances of algorithms were iteratively trained and validated on data from eight centers and tested on the ninth for both scenarios separately. Segmentation performance was assessed using the Dice similarity coefficient (DSC), while classification performance was assessed using metrics, such as precision, recall, F1-score, accuracy, area under the Receiver Operating Characteristic (ROC AUC), and area under the precision-recall curve (PRC AUC). ResUNet achieved DSC values of 0.84$\pm$0.03, 0.71$\pm$0.06, and 0.86$\pm$0.02 for MNG, TH, and DG, respectively. Classification in scenario 1 achieved an accuracy of 0.76$\pm$0.04 and a ROC AUC of 0.92$\pm$0.02 while in scenario 2, classification yielded an accuracy of 0.74$\pm$0.05 and a ROC AUC of 0.90$\pm$0.02. The automated pipeline demonstrated comparable performance to physician segmentations on several classification metrics across different classes, effectively reducing assessment time while maintaining high diagnostic accuracy. Code available at: https://github.com/ahxmeds/thyroidiomics.git.
Deep learning-based attenuation correction in the image domain for myocardial perfusion SPECT imaging
Feb 10, 2021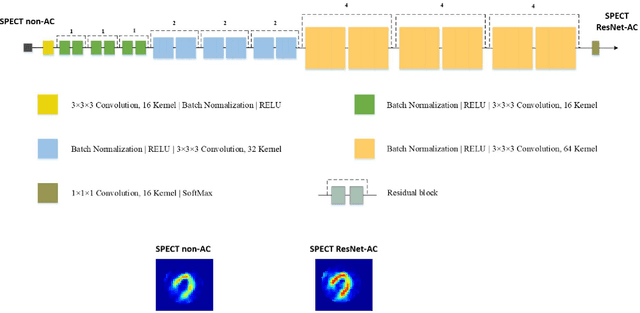
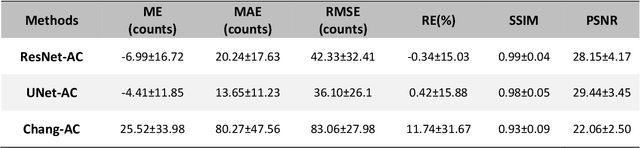
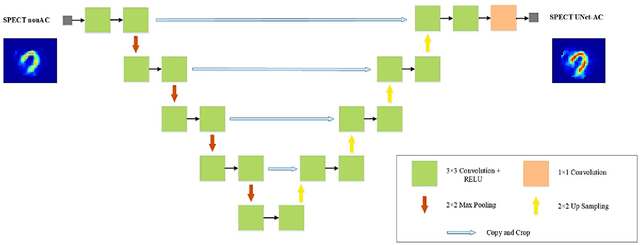
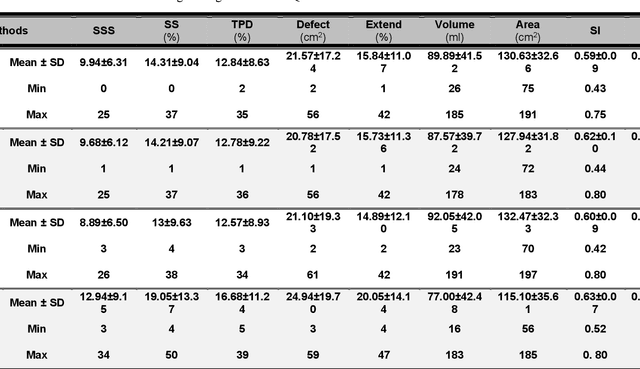
Abstract:Objective: In this work, we set out to investigate the accuracy of direct attenuation correction (AC) in the image domain for the myocardial perfusion SPECT imaging (MPI-SPECT) using two residual (ResNet) and UNet deep convolutional neural networks. Methods: The MPI-SPECT 99mTc-sestamibi images of 99 participants were retrospectively examined. UNet and ResNet networks were trained using SPECT non-attenuation corrected images as input and CT-based attenuation corrected SPECT images (CT-AC) as reference. The Chang AC approach, considering a uniform attenuation coefficient within the body contour, was also implemented. Quantitative and clinical evaluation of the proposed methods were performed considering SPECT CT-AC images of 19 subjects as reference using the mean absolute error (MAE), structural similarity index (SSIM) metrics, as well as relevant clinical indices such as perfusion deficit (TPD). Results: Overall, the deep learning solution exhibited good agreement with the CT-based AC, noticeably outperforming the Chang method. The ResNet and UNet models resulted in the ME (count) of ${-6.99\pm16.72}$ and ${-4.41\pm11.8}$ and SSIM of ${0.99\pm0.04}$ and ${0.98\pm0.05}$, respectively. While the Change approach led to ME and SSIM of ${25.52\pm33.98}$ and ${0.93\pm0.09}$, respectively. Similarly, the clinical evaluation revealed a mean TPD of ${12.78\pm9.22}$ and ${12.57\pm8.93}$ for the ResNet and UNet models, respectively, compared to ${12.84\pm8.63}$ obtained from the reference SPECT CT-AC images. On the other hand, the Chang approach led to a mean TPD of ${16.68\pm11.24}$. Conclusion: We evaluated two deep convolutional neural networks to estimate SPECT-AC images directly from the non-attenuation corrected images. The deep learning solutions exhibited the promising potential to generate reliable attenuation corrected SPECT images without the use of transmission scanning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge