Jean-Francois Rajotte
Observer study-based evaluation of TGAN architecture used to generate oncological PET images
Nov 28, 2023Abstract:The application of computer-vision algorithms in medical imaging has increased rapidly in recent years. However, algorithm training is challenging due to limited sample sizes, lack of labeled samples, as well as privacy concerns regarding data sharing. To address these issues, we previously developed (Bergen et al. 2022) a synthetic PET dataset for Head and Neck (H and N) cancer using the temporal generative adversarial network (TGAN) architecture and evaluated its performance segmenting lesions and identifying radiomics features in synthesized images. In this work, a two-alternative forced-choice (2AFC) observer study was performed to quantitatively evaluate the ability of human observers to distinguish between real and synthesized oncological PET images. In the study eight trained readers, including two board-certified nuclear medicine physicians, read 170 real/synthetic image pairs presented as 2D-transaxial using a dedicated web app. For each image pair, the observer was asked to identify the real image and input their confidence level with a 5-point Likert scale. P-values were computed using the binomial test and Wilcoxon signed-rank test. A heat map was used to compare the response accuracy distribution for the signed-rank test. Response accuracy for all observers ranged from 36.2% [27.9-44.4] to 63.1% [54.8-71.3]. Six out of eight observers did not identify the real image with statistical significance, indicating that the synthetic dataset was reasonably representative of oncological PET images. Overall, this study adds validity to the realism of our simulated H&N cancer dataset, which may be implemented in the future to train AI algorithms while favoring patient confidentiality and privacy protection.
Assessing Privacy Leakage in Synthetic 3-D PET Imaging using Transversal GAN
Jun 13, 2022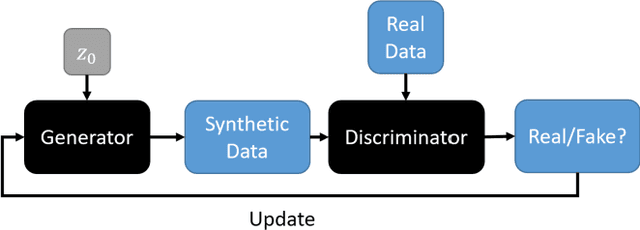
Abstract:Training computer-vision related algorithms on medical images for disease diagnosis or image segmentation is difficult in large part due to privacy concerns. For this reason, generative image models are highly sought after to facilitate data sharing. However, 3-D generative models are understudied, and investigation of their privacy leakage is needed. We introduce our 3-D generative model, Transversal GAN (TrGAN), using head & neck PET images which are conditioned on tumour masks as a case study. We define quantitative measures of image fidelity, utility and privacy for our model. These metrics are evaluated in the course of training to identify ideal fidelity, utility and privacy trade-offs and establish the relationships between these parameters. We show that the discriminator of the TrGAN is vulnerable to attack, and that an attacker can identify which samples were used in training with almost perfect accuracy (AUC = 0.99). We also show that an attacker with access to only the generator cannot reliably classify whether a sample had been used for training (AUC = 0.51). This suggests that TrGAN generators, but not discriminators, may be used for sharing synthetic 3-D PET data with minimal privacy risk while maintaining good utility and fidelity.
Group GAN
May 27, 2022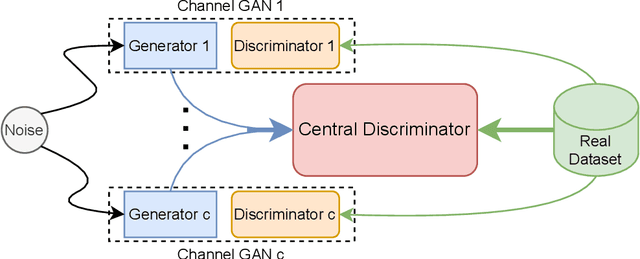



Abstract:Generating multivariate time series is a promising approach for sharing sensitive data in many medical, financial, and IoT applications. A common type of multivariate time series originates from a single source such as the biometric measurements from a medical patient. This leads to complex dynamical patterns between individual time series that are hard to learn by typical generation models such as GANs. There is valuable information in those patterns that machine learning models can use to better classify, predict or perform other downstream tasks. We propose a novel framework that takes time series' common origin into account and favors inter-channel relationship preservation. The two key points of our method are: 1) the individual time series are generated from a common point in latent space and 2) a central discriminator favors the preservation of inter-channel dynamics. We demonstrate empirically that our method helps preserve channel correlations and that our synthetic data performs very well downstream tasks with medical and financial data.
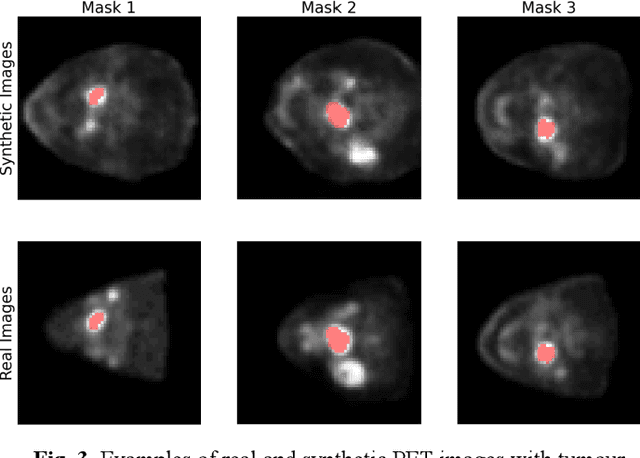
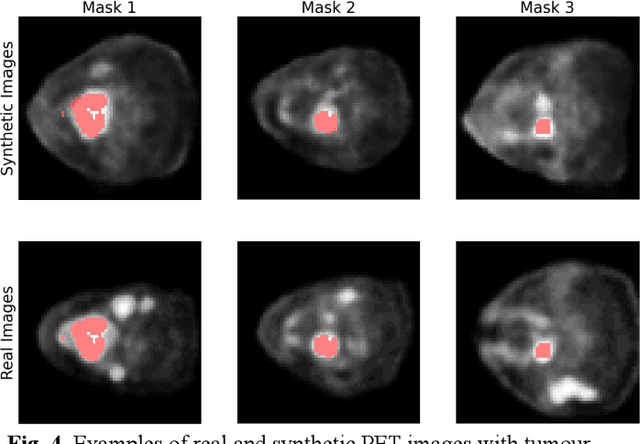
3-D PET Image Generation with tumour masks using TGAN
Nov 02, 2021



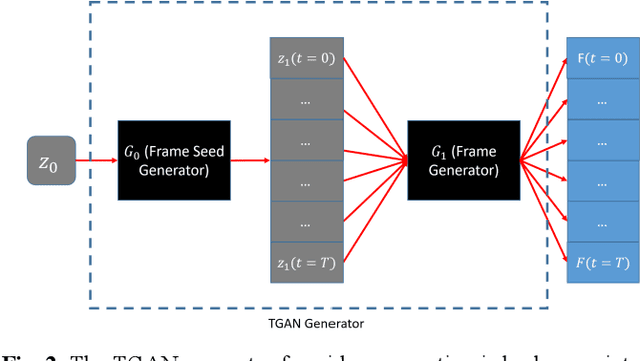
Abstract:Training computer-vision related algorithms on medical images for disease diagnosis or image segmentation is difficult due to the lack of training data, labeled samples, and privacy concerns. For this reason, a robust generative method to create synthetic data is highly sought after. However, most three-dimensional image generators require additional image input or are extremely memory intensive. To address these issues we propose adapting video generation techniques for 3-D image generation. Using the temporal GAN (TGAN) architecture, we show we are able to generate realistic head and neck PET images. We also show that by conditioning the generator on tumour masks, we are able to control the geometry and location of the tumour in the generated images. To test the utility of the synthetic images, we train a segmentation model using the synthetic images. Synthetic images conditioned on real tumour masks are automatically segmented, and the corresponding real images are also segmented. We evaluate the segmentations using the Dice score and find the segmentation algorithm performs similarly on both datasets (0.65 synthetic data, 0.70 real data). Various radionomic features are then calculated over the segmented tumour volumes for each data set. A comparison of the real and synthetic feature distributions show that seven of eight feature distributions had statistically insignificant differences (p>0.05). Correlation coefficients were also calculated between all radionomic features and it is shown that all of the strong statistical correlations in the real data set are preserved in the synthetic data set.
Reducing bias and increasing utility by federated generative modeling of medical images using a centralized adversary
Jan 18, 2021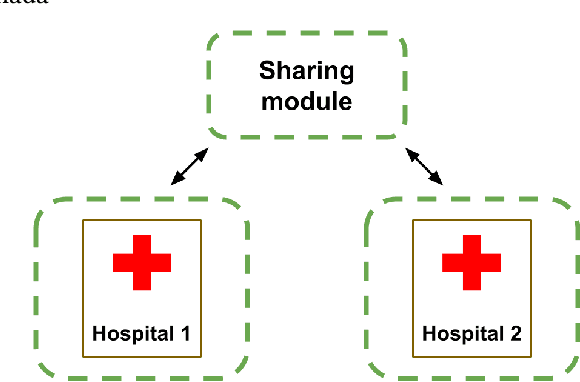

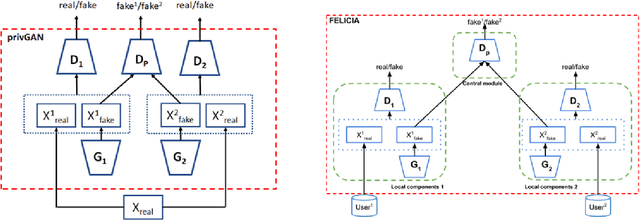

Abstract:We introduce FELICIA (FEderated LearnIng with a CentralIzed Adversary) a generative mechanism enabling collaborative learning. In particular, we show how a data owner with limited and biased data could benefit from other data owners while keeping data from all the sources private. This is a common scenario in medical image analysis where privacy legislation prevents data from being shared outside local premises. FELICIA works for a large family of Generative Adversarial Networks (GAN) architectures including vanilla and conditional GANs as demonstrated in this work. We show that by using the FELICIA mechanism, a data owner with limited image samples can generate high-quality synthetic images with high utility while neither data owners has to provide access to its data. The sharing happens solely through a central discriminator that has access limited to synthetic data. Here, utility is defined as classification performance on a real test set. We demonstrate these benefits on several realistic healthcare scenarions using benchmark image datasets (MNIST, CIFAR-10) as well as on medical images for the task of skin lesion classification. With multiple experiments, we show that even in the worst cases, combining FELICIA with real data gracefully achieves performance on par with real data while most results significantly improves the utility.
Private data sharing between decentralized users through the privGAN architecture
Sep 14, 2020



Abstract:More data is almost always beneficial for analysis and machine learning tasks. In many realistic situations however, an enterprise cannot share its data, either to keep a competitive advantage or to protect the privacy of the data sources, the enterprise's clients for example. We propose a method for data owners to share synthetic or fake versions of their data without sharing the actual data, nor the parameters of models that have direct access to the data. The method proposed is based on the privGAN architecture where local GANs are trained on their respective data subsets with an extra penalty from a central discriminator aiming to discriminate the origin of a given fake sample. We demonstrate that this approach, when applied to subsets of various sizes, leads to better utility for the owners than the utility from their real small datasets. The only shared pieces of information are the parameter updates of the central discriminator. The privacy is demonstrated with white-box attacks on the most vulnerable elments of the architecture and the results are close to random guessing. This method would apply naturally in a federated learning setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge