Yijia Zheng
Using Large Language Models to Tackle Fundamental Challenges in Graph Learning: A Comprehensive Survey
May 24, 2025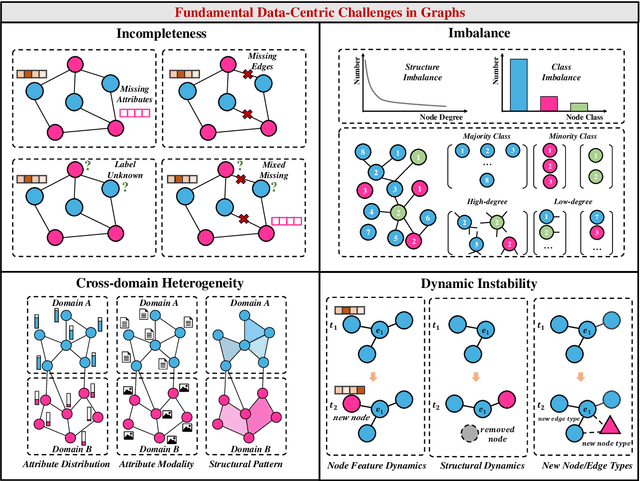
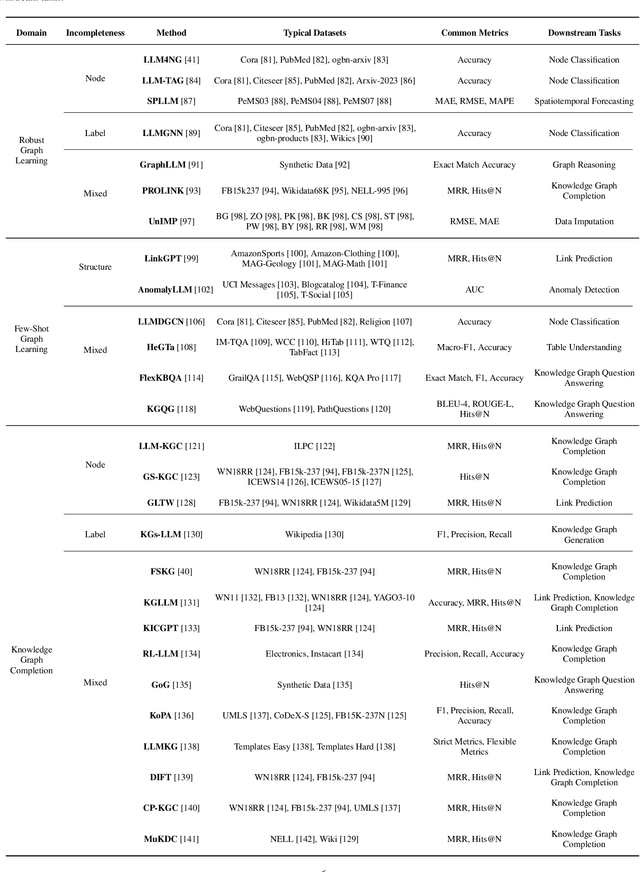
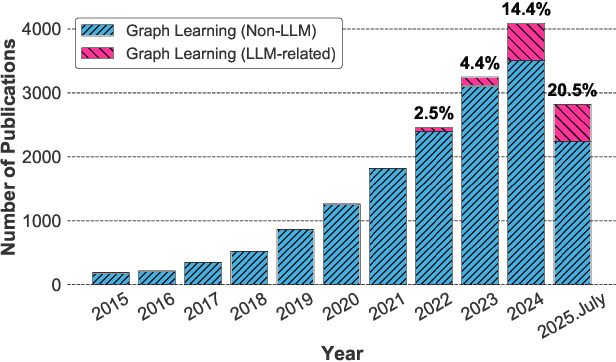
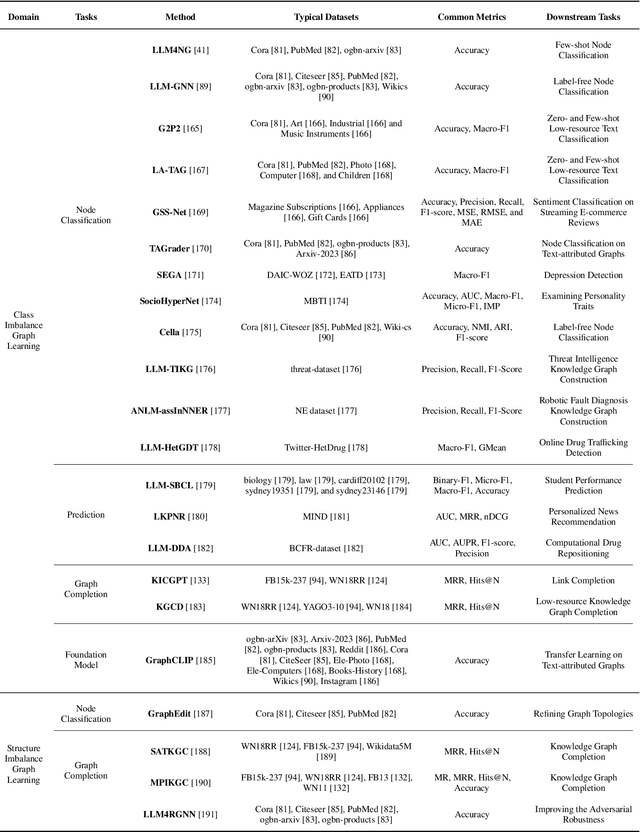
Abstract:Graphs are a widely used paradigm for representing non-Euclidean data, with applications ranging from social network analysis to biomolecular prediction. Conventional graph learning approaches typically rely on fixed structural assumptions or fully observed data, limiting their effectiveness in more complex, noisy, or evolving settings. Consequently, real-world graph data often violates the assumptions of traditional graph learning methods, in particular, it leads to four fundamental challenges: (1) Incompleteness, real-world graphs have missing nodes, edges, or attributes; (2) Imbalance, the distribution of the labels of nodes or edges and their structures for real-world graphs are highly skewed; (3) Cross-domain Heterogeneity, graphs from different domains exhibit incompatible feature spaces or structural patterns; and (4) Dynamic Instability, graphs evolve over time in unpredictable ways. Recent advances in Large Language Models (LLMs) offer the potential to tackle these challenges by leveraging rich semantic reasoning and external knowledge. This survey provides a comprehensive review of how LLMs can be integrated with graph learning to address the aforementioned challenges. For each challenge, we review both traditional solutions and modern LLM-driven approaches, highlighting how LLMs contribute unique advantages. Finally, we discuss open research questions and promising future directions in this emerging interdisciplinary field. To support further exploration, we have curated a repository of recent advances on graph learning challenges: https://github.com/limengran98/Awesome-Literature-Graph-Learning-Challenges.
Co-Representation Neural Hypergraph Diffusion for Edge-Dependent Node Classification
May 23, 2024Abstract:Hypergraphs are widely employed to represent complex higher-order relationships in real-world applications. Most hypergraph learning research focuses on node- or edge-level tasks. A practically relevant but more challenging task, edge-dependent node classification (ENC), is only recently proposed. In ENC, a node can have different labels across different hyperedges, which requires the modeling of node-hyperedge pairs instead of single nodes or hyperedges. Existing solutions for this task are based on message passing and model within-edge and within-node interactions as multi-input single-output functions. This brings three limitations: (1) non-adaptive representation size, (2) node/edge agnostic messages, and (3) insufficient interactions among nodes or hyperedges. To tackle these limitations, we develop CoNHD, a new solution based on hypergraph diffusion. Specifically, we first extend hypergraph diffusion using node-hyperedge co-representations. This extension explicitly models both within-edge and within-node interactions as multi-input multi-output functions using two equivariant diffusion operators. To avoid handcrafted regularization functions, we propose a neural implementation for the co-representation hypergraph diffusion process. Extensive experiments demonstrate the effectiveness and efficiency of the proposed CoNHD model.
IMMA: Immunizing text-to-image Models against Malicious Adaptation
Nov 30, 2023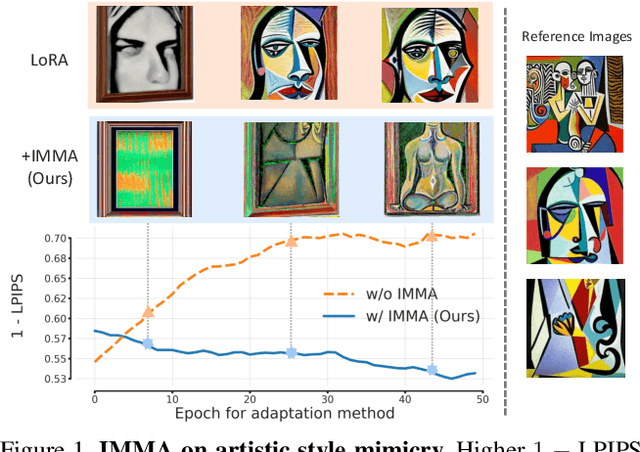
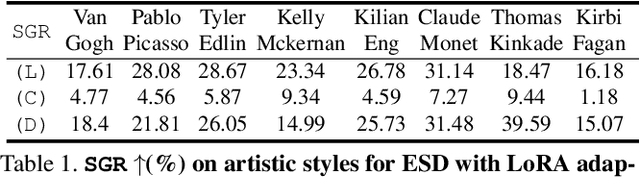


Abstract:Advancements in text-to-image models and fine-tuning methods have led to the increasing risk of malicious adaptation, i.e., fine-tuning to generate harmful unauthorized content. Recent works, e.g., Glaze or MIST, have developed data-poisoning techniques which protect the data against adaptation methods. In this work, we consider an alternative paradigm for protection. We propose to ``immunize'' the model by learning model parameters that are difficult for the adaptation methods when fine-tuning malicious content; in short IMMA. Empirical results show IMMA's effectiveness against malicious adaptations, including mimicking the artistic style and learning of inappropriate/unauthorized content, over three adaptation methods: LoRA, Textual-Inversion, and DreamBooth.
Learning Manifold Dimensions with Conditional Variational Autoencoders
Feb 23, 2023Abstract:Although the variational autoencoder (VAE) and its conditional extension (CVAE) are capable of state-of-the-art results across multiple domains, their precise behavior is still not fully understood, particularly in the context of data (like images) that lie on or near a low-dimensional manifold. For example, while prior work has suggested that the globally optimal VAE solution can learn the correct manifold dimension, a necessary (but not sufficient) condition for producing samples from the true data distribution, this has never been rigorously proven. Moreover, it remains unclear how such considerations would change when various types of conditioning variables are introduced, or when the data support is extended to a union of manifolds (e.g., as is likely the case for MNIST digits and related). In this work, we address these points by first proving that VAE global minima are indeed capable of recovering the correct manifold dimension. We then extend this result to more general CVAEs, demonstrating practical scenarios whereby the conditioning variables allow the model to adaptively learn manifolds of varying dimension across samples. Our analyses, which have practical implications for various CVAE design choices, are also supported by numerical results on both synthetic and real-world datasets.
Graph Polish: A Novel Graph Generation Paradigm for Molecular Optimization
Aug 14, 2020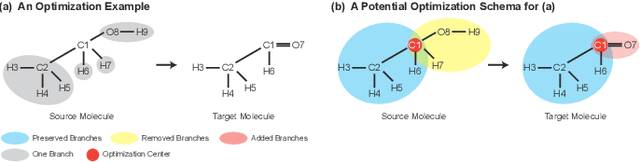
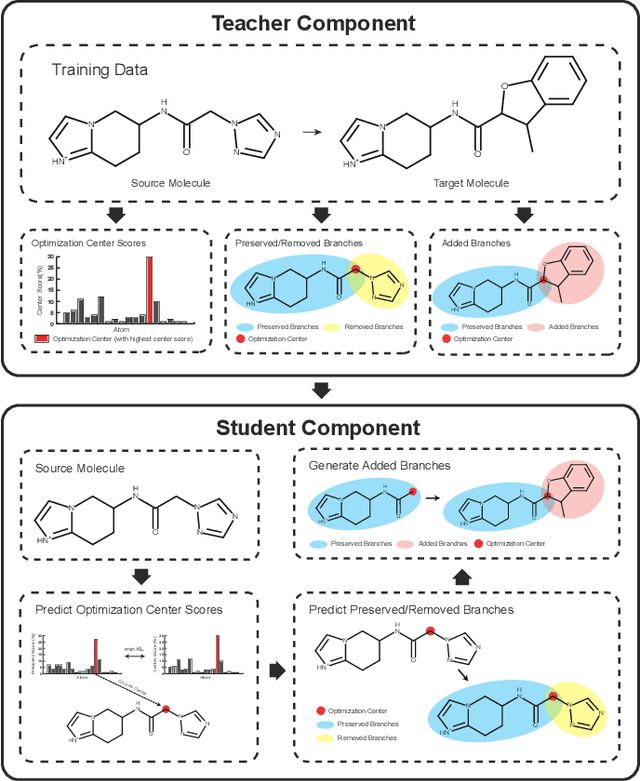
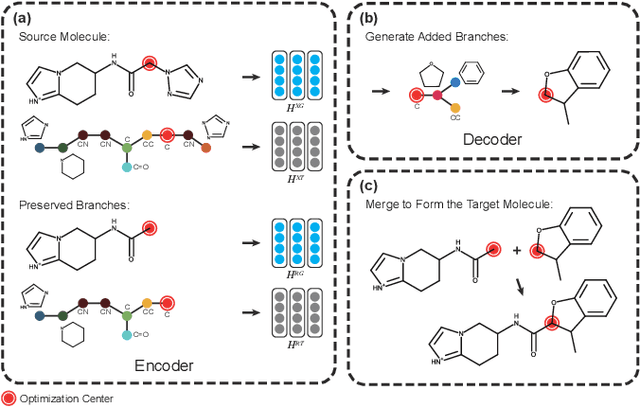
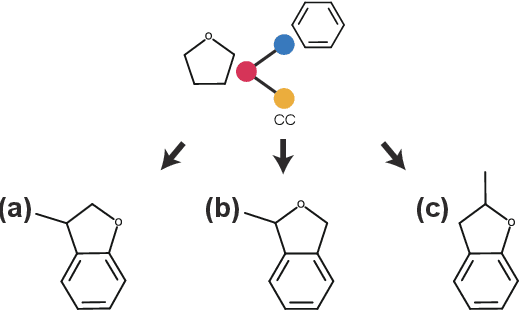
Abstract:Molecular optimization, which transforms a given input molecule X into another Y with desirable properties, is essential in molecular drug discovery. The traditional translating approaches, generating the molecular graphs from scratch by adding some substructures piece by piece, prone to error because of the large set of candidate substructures in a large number of steps to the final target. In this study, we present a novel molecular optimization paradigm, Graph Polish, which changes molecular optimization from the traditional "two-language translating" task into a "single-language polishing" task. The key to this optimization paradigm is to find an optimization center subject to the conditions that the preserved areas around it ought to be maximized and thereafter the removed and added regions should be minimized. We then propose an effective and efficient learning framework T&S polish to capture the long-term dependencies in the optimization steps. The T component automatically identifies and annotates the optimization centers and the preservation, removal and addition of some parts of the molecule, and the S component learns these behaviors and applies these actions to a new molecule. Furthermore, the proposed paradigm can offer an intuitive interpretation for each molecular optimization result. Experiments with multiple optimization tasks are conducted on four benchmark datasets. The proposed T&S polish approach achieves significant advantage over the five state-of-the-art baseline methods on all the tasks. In addition, extensive studies are conducted to validate the effectiveness, explainability and time saving of the novel optimization paradigm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge