Yunpeng Cai
Automated Learning of Semantic Embedding Representations for Diffusion Models
May 09, 2025Abstract:Generative models capture the true distribution of data, yielding semantically rich representations. Denoising diffusion models (DDMs) exhibit superior generative capabilities, though efficient representation learning for them are lacking. In this work, we employ a multi-level denoising autoencoder framework to expand the representation capacity of DDMs, which introduces sequentially consistent Diffusion Transformers and an additional timestep-dependent encoder to acquire embedding representations on the denoising Markov chain through self-conditional diffusion learning. Intuitively, the encoder, conditioned on the entire diffusion process, compresses high-dimensional data into directional vectors in latent under different noise levels, facilitating the learning of image embeddings across all timesteps. To verify the semantic adequacy of embeddings generated through this approach, extensive experiments are conducted on various datasets, demonstrating that optimally learned embeddings by DDMs surpass state-of-the-art self-supervised representation learning methods in most cases, achieving remarkable discriminative semantic representation quality. Our work justifies that DDMs are not only suitable for generative tasks, but also potentially advantageous for general-purpose deep learning applications.
A Weakly Supervised and Globally Explainable Learning Framework for Brain Tumor Segmentation
Aug 02, 2024



Abstract:Machine-based brain tumor segmentation can help doctors make better diagnoses. However, the complex structure of brain tumors and expensive pixel-level annotations present challenges for automatic tumor segmentation. In this paper, we propose a counterfactual generation framework that not only achieves exceptional brain tumor segmentation performance without the need for pixel-level annotations, but also provides explainability. Our framework effectively separates class-related features from class-unrelated features of the samples, and generate new samples that preserve identity features while altering class attributes by embedding different class-related features. We perform topological data analysis on the extracted class-related features and obtain a globally explainable manifold, and for each abnormal sample to be segmented, a meaningful normal sample could be effectively generated with the guidance of the rule-based paths designed within the manifold for comparison for identifying the tumor regions. We evaluate our proposed method on two datasets, which demonstrates superior performance of brain tumor segmentation. The code is available at https://github.com/xrt11/tumor-segmentation.
Accurate Explanation Model for Image Classifiers using Class Association Embedding
Jun 12, 2024Abstract:Image classification is a primary task in data analysis where explainable models are crucially demanded in various applications. Although amounts of methods have been proposed to obtain explainable knowledge from the black-box classifiers, these approaches lack the efficiency of extracting global knowledge regarding the classification task, thus is vulnerable to local traps and often leads to poor accuracy. In this study, we propose a generative explanation model that combines the advantages of global and local knowledge for explaining image classifiers. We develop a representation learning method called class association embedding (CAE), which encodes each sample into a pair of separated class-associated and individual codes. Recombining the individual code of a given sample with altered class-associated code leads to a synthetic real-looking sample with preserved individual characters but modified class-associated features and possibly flipped class assignments. A building-block coherency feature extraction algorithm is proposed that efficiently separates class-associated features from individual ones. The extracted feature space forms a low-dimensional manifold that visualizes the classification decision patterns. Explanation on each individual sample can be then achieved in a counter-factual generation manner which continuously modifies the sample in one direction, by shifting its class-associated code along a guided path, until its classification outcome is changed. We compare our method with state-of-the-art ones on explaining image classification tasks in the form of saliency maps, demonstrating that our method achieves higher accuracies. The code is available at https://github.com/xrt11/XAI-CODE.
Deep Learning Method to Predict Wound Healing Progress Based on Collagen Fibers in Wound Tissue
May 08, 2024Abstract:Wound healing is a complex process involving changes in collagen fibers. Accurate monitoring of these changes is crucial for assessing the progress of wound healing and has significant implications for guiding clinical treatment strategies and drug screening. However, traditional quantitative analysis methods focus on spatial characteristics such as collagen fiber alignment and variance, lacking threshold standards to differentiate between different stages of wound healing. To address this issue, we propose an innovative approach based on deep learning to predict the progression of wound healing by analyzing collagen fiber features in histological images of wound tissue. Leveraging the unique learning capabilities of deep learning models, our approach captures the feature variations of collagen fibers in histological images from different categories and classifies them into various stages of wound healing. To overcome the limited availability of histological image data, we employ a transfer learning strategy. Specifically, we fine-tune a VGG16 model pretrained on the ImageNet dataset to adapt it to the classification task of histological images of wounds. Through this process, our model achieves 82% accuracy in classifying six stages of wound healing. Furthermore, to enhance the interpretability of the model, we employ a class activation mapping technique called LayerCAM. LayerCAM reveals the image regions on which the model relies when making predictions, providing transparency to the model's decision-making process. This visualization not only helps us understand how the model identifies and evaluates collagen fiber features but also enhances trust in the model's prediction results. To the best of our knowledge, our proposed model is the first deep learning-based classification model used for predicting wound healing stages.
Active Globally Explainable Learning for Medical Images via Class Association Embedding and Cyclic Adversarial Generation
Jun 12, 2023Abstract:Explainability poses a major challenge to artificial intelligence (AI) techniques. Current studies on explainable AI (XAI) lack the efficiency of extracting global knowledge about the learning task, thus suffer deficiencies such as imprecise saliency, context-aware absence and vague meaning. In this paper, we propose the class association embedding (CAE) approach to address these issues. We employ an encoder-decoder architecture to embed sample features and separate them into class-related and individual-related style vectors simultaneously. Recombining the individual-style code of a given sample with the class-style code of another leads to a synthetic sample with preserved individual characters but changed class assignment, following a cyclic adversarial learning strategy. Class association embedding distills the global class-related features of all instances into a unified domain with well separation between classes. The transition rules between different classes can be then extracted and further employed to individual instances. We then propose an active XAI framework which manipulates the class-style vector of a certain sample along guided paths towards the counter-classes, resulting in a series of counter-example synthetic samples with identical individual characters. Comparing these counterfactual samples with the original ones provides a global, intuitive illustration to the nature of the classification tasks. We adopt the framework on medical image classification tasks, which show that more precise saliency maps with powerful context-aware representation can be achieved compared with existing methods. Moreover, the disease pathology can be directly visualized via traversing the paths in the class-style space.
Smoothness Sensor: Adaptive Smoothness-Transition Graph Convolutions for Attributed Graph Clustering
Sep 12, 2020Abstract:Clustering techniques attempt to group objects with similar properties into a cluster. Clustering the nodes of an attributed graph, in which each node is associated with a set of feature attributes, has attracted significant attention. Graph convolutional networks (GCNs) represent an effective approach for integrating the two complementary factors of node attributes and structural information for attributed graph clustering. However, oversmoothing of GCNs produces indistinguishable representations of nodes, such that the nodes in a graph tend to be grouped into fewer clusters, and poses a challenge due to the resulting performance drop. In this study, we propose a smoothness sensor for attributed graph clustering based on adaptive smoothness-transition graph convolutions, which senses the smoothness of a graph and adaptively terminates the current convolution once the smoothness is saturated to prevent oversmoothing. Furthermore, as an alternative to graph-level smoothness, a novel fine-gained node-wise level assessment of smoothness is proposed, in which smoothness is computed in accordance with the neighborhood conditions of a given node at a certain order of graph convolution. In addition, a self-supervision criterion is designed considering both the tightness within clusters and the separation between clusters to guide the whole neural network training process. Experiments show that the proposed methods significantly outperform 12 other state-of-the-art baselines in terms of three different metrics across four benchmark datasets. In addition, an extensive study reveals the reasons for their effectiveness and efficiency.
Graph Polish: A Novel Graph Generation Paradigm for Molecular Optimization
Aug 14, 2020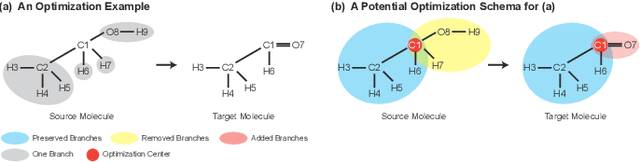
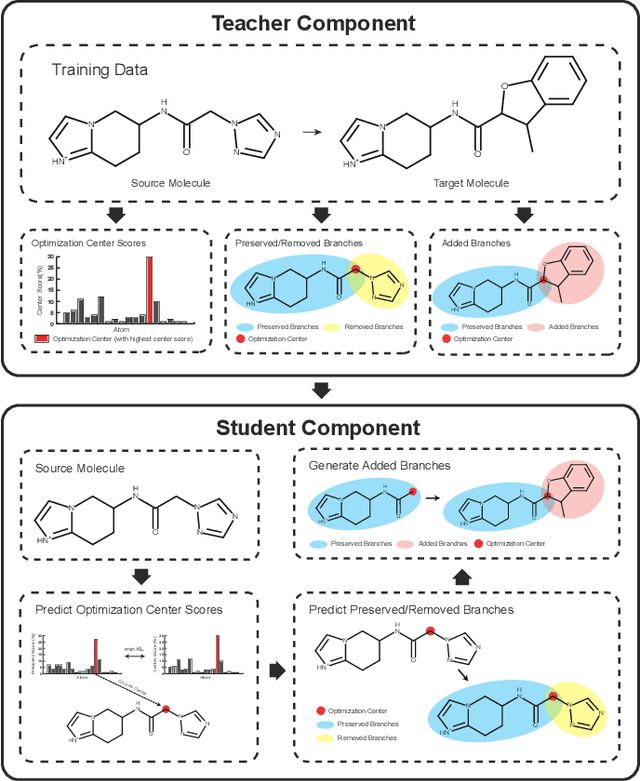
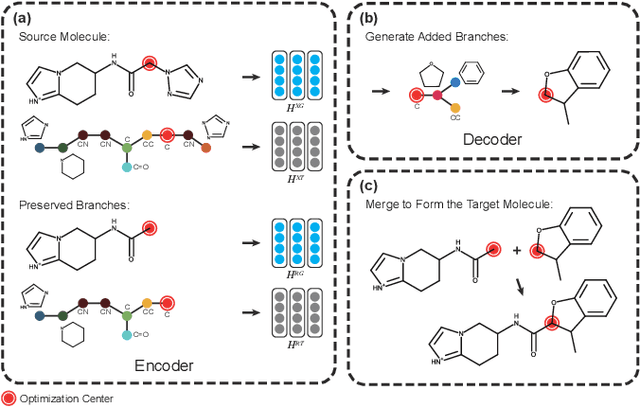
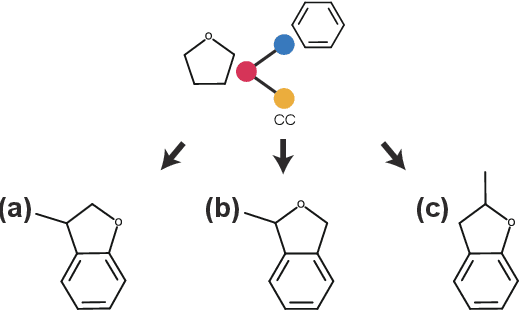
Abstract:Molecular optimization, which transforms a given input molecule X into another Y with desirable properties, is essential in molecular drug discovery. The traditional translating approaches, generating the molecular graphs from scratch by adding some substructures piece by piece, prone to error because of the large set of candidate substructures in a large number of steps to the final target. In this study, we present a novel molecular optimization paradigm, Graph Polish, which changes molecular optimization from the traditional "two-language translating" task into a "single-language polishing" task. The key to this optimization paradigm is to find an optimization center subject to the conditions that the preserved areas around it ought to be maximized and thereafter the removed and added regions should be minimized. We then propose an effective and efficient learning framework T&S polish to capture the long-term dependencies in the optimization steps. The T component automatically identifies and annotates the optimization centers and the preservation, removal and addition of some parts of the molecule, and the S component learns these behaviors and applies these actions to a new molecule. Furthermore, the proposed paradigm can offer an intuitive interpretation for each molecular optimization result. Experiments with multiple optimization tasks are conducted on four benchmark datasets. The proposed T&S polish approach achieves significant advantage over the five state-of-the-art baseline methods on all the tasks. In addition, extensive studies are conducted to validate the effectiveness, explainability and time saving of the novel optimization paradigm.
HopGAT: Hop-aware Supervision Graph Attention Networks for Sparsely Labeled Graphs
Apr 09, 2020
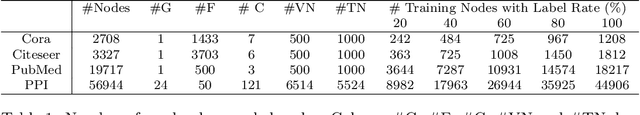
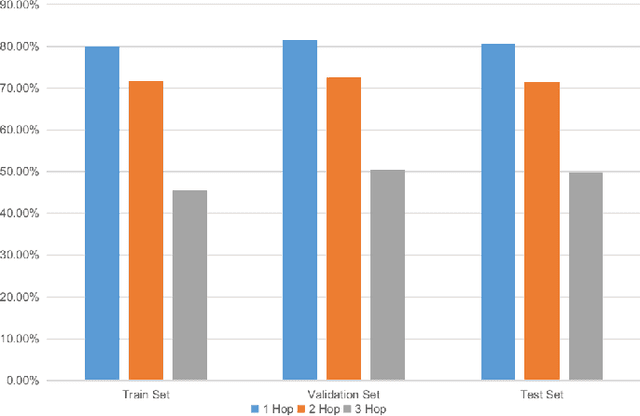

Abstract:Due to the cost of labeling nodes, classifying a node in a sparsely labeled graph while maintaining the prediction accuracy deserves attention. The key point is how the algorithm learns sufficient information from more neighbors with different hop distances. This study first proposes a hop-aware attention supervision mechanism for the node classification task. A simulated annealing learning strategy is then adopted to balance two learning tasks, node classification and the hop-aware attention coefficients, along the training timeline. Compared with state-of-the-art models, the experimental results proved the superior effectiveness of the proposed Hop-aware Supervision Graph Attention Networks (HopGAT) model. Especially, for the protein-protein interaction network, in a 40% labeled graph, the performance loss is only 3.9%, from 98.5% to 94.6%, compared to the fully labeled graph. Extensive experiments also demonstrate the effectiveness of supervised attention coefficient and learning strategies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge