Rui Xiao
Training-free Uncertainty Guidance for Complex Visual Tasks with MLLMs
Oct 01, 2025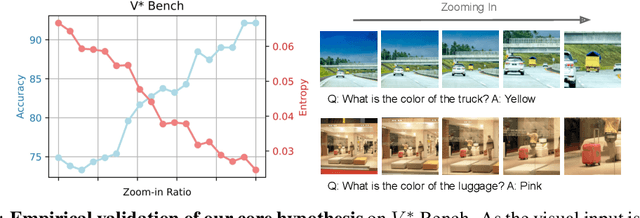
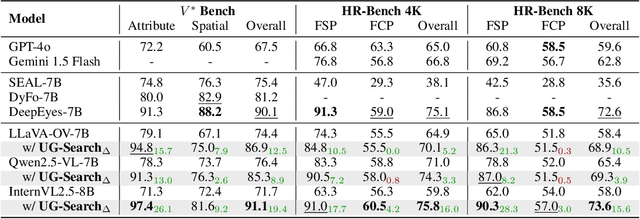
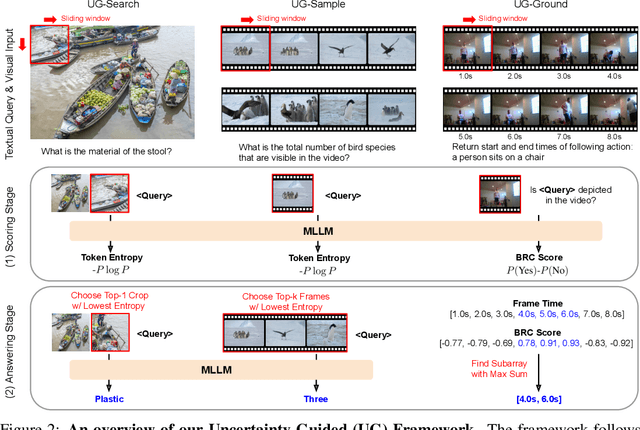
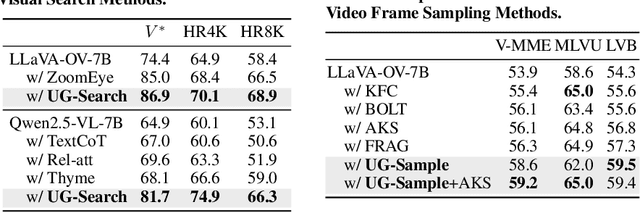
Abstract:Multimodal Large Language Models (MLLMs) often struggle with fine-grained perception, such as identifying small objects in high-resolution images or finding key moments in long videos. Existing works typically rely on complicated, task-specific fine-tuning, which limits their generalizability and increases model complexity. In this work, we propose an effective, training-free framework that uses an MLLM's intrinsic uncertainty as a proactive guidance signal. Our core insight is that a model's output entropy decreases when presented with relevant visual information. We introduce a unified mechanism that scores candidate visual inputs by response uncertainty, enabling the model to autonomously focus on the most salient data. We apply this simple principle to three complex visual tasks: Visual Search, Long Video Understanding, and Temporal Grounding, allowing off-the-shelf MLLMs to achieve performance competitive with specialized, fine-tuned methods. Our work validates that harnessing intrinsic uncertainty is a powerful, general strategy for enhancing fine-grained multimodal performance.
Conformable Convolution for Topologically Aware Learning of Complex Anatomical Structures
Dec 29, 2024Abstract:While conventional computer vision emphasizes pixel-level and feature-based objectives, medical image analysis of intricate biological structures necessitates explicit representation of their complex topological properties. Despite their successes, deep learning models often struggle to accurately capture the connectivity and continuity of fine, sometimes pixel-thin, yet critical structures due to their reliance on implicit learning from data. Such shortcomings can significantly impact the reliability of analysis results and hinder clinical decision-making. To address this challenge, we introduce Conformable Convolution, a novel convolutional layer designed to explicitly enforce topological consistency. Conformable Convolution learns adaptive kernel offsets that preferentially focus on regions of high topological significance within an image. This prioritization is guided by our proposed Topological Posterior Generator (TPG) module, which leverages persistent homology. The TPG module identifies key topological features and guides the convolutional layers by applying persistent homology to feature maps transformed into cubical complexes. Our proposed modules are architecture-agnostic, enabling them to be integrated seamlessly into various architectures. We showcase the effectiveness of our framework in the segmentation task, where preserving the interconnectedness of structures is critical. Experimental results on three diverse datasets demonstrate that our framework effectively preserves the topology in the segmentation downstream task, both quantitatively and qualitatively.
Molly: Making Large Language Model Agents Solve Python Problem More Logically
Dec 24, 2024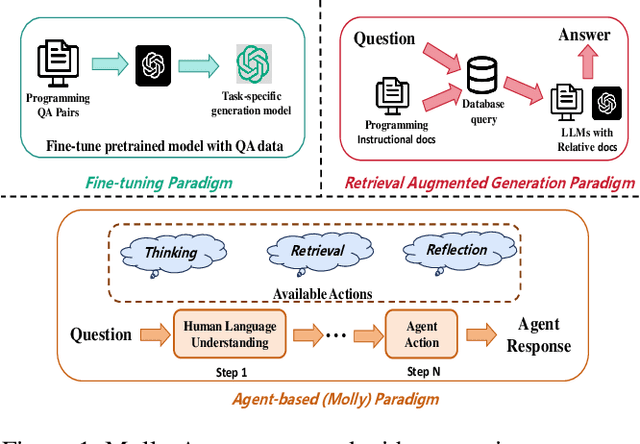
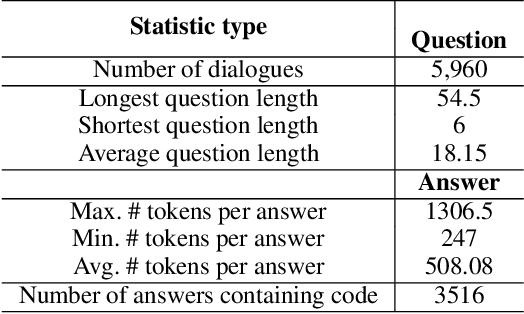
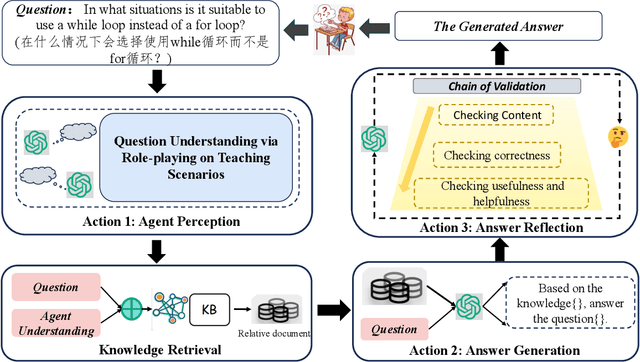
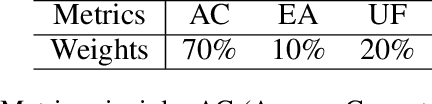
Abstract:Applying large language models (LLMs) as teaching assists has attracted much attention as an integral part of intelligent education, particularly in computing courses. To reduce the gap between the LLMs and the computer programming education expert, fine-tuning and retrieval augmented generation (RAG) are the two mainstream methods in existing researches. However, fine-tuning for specific tasks is resource-intensive and may diminish the model`s generalization capabilities. RAG can perform well on reducing the illusion of LLMs, but the generation of irrelevant factual content during reasoning can cause significant confusion for learners. To address these problems, we introduce the Molly agent, focusing on solving the proposed problem encountered by learners when learning Python programming language. Our agent automatically parse the learners' questioning intent through a scenario-based interaction, enabling precise retrieval of relevant documents from the constructed knowledge base. At generation stage, the agent reflect on the generated responses to ensure that they not only align with factual content but also effectively answer the user's queries. Extensive experimentation on a constructed Chinese Python QA dataset shows the effectiveness of the Molly agent, indicating an enhancement in its performance for providing useful responses to Python questions.
FLAIR: VLM with Fine-grained Language-informed Image Representations
Dec 04, 2024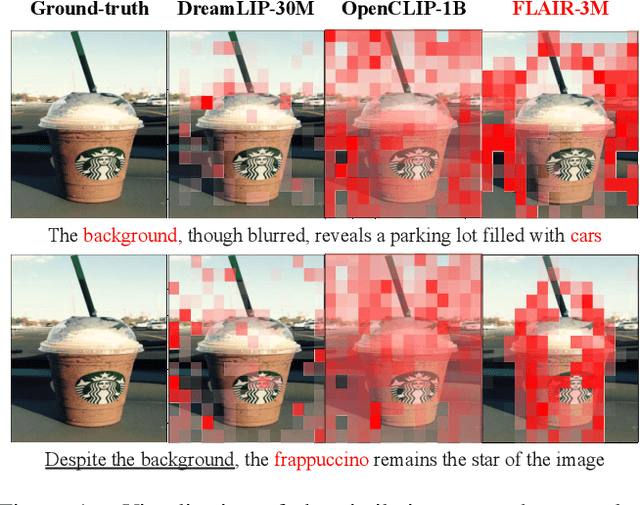
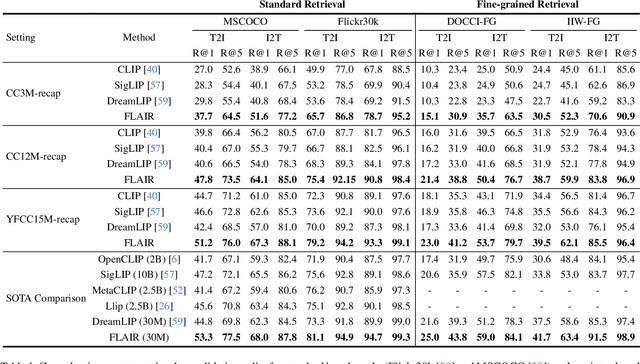
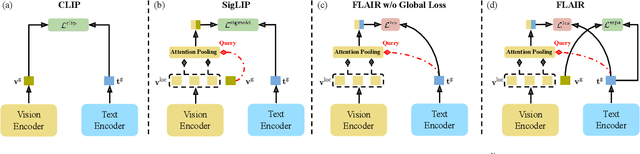
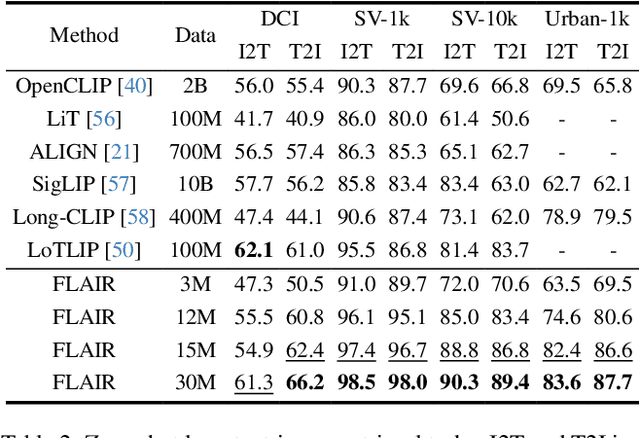
Abstract:CLIP has shown impressive results in aligning images and texts at scale. However, its ability to capture detailed visual features remains limited because CLIP matches images and texts at a global level. To address this issue, we propose FLAIR, Fine-grained Language-informed Image Representations, an approach that utilizes long and detailed image descriptions to learn localized image embeddings. By sampling diverse sub-captions that describe fine-grained details about an image, we train our vision-language model to produce not only global embeddings but also text-specific image representations. Our model introduces text-conditioned attention pooling on top of local image tokens to produce fine-grained image representations that excel at retrieving detailed image content. We achieve state-of-the-art performance on both, existing multimodal retrieval benchmarks, as well as, our newly introduced fine-grained retrieval task which evaluates vision-language models' ability to retrieve partial image content. Furthermore, our experiments demonstrate the effectiveness of FLAIR trained on 30M image-text pairs in capturing fine-grained visual information, including zero-shot semantic segmentation, outperforming models trained on billions of pairs. Code is available at https://github.com/ExplainableML/flair .
COSMOS: Cross-Modality Self-Distillation for Vision Language Pre-training
Dec 02, 2024
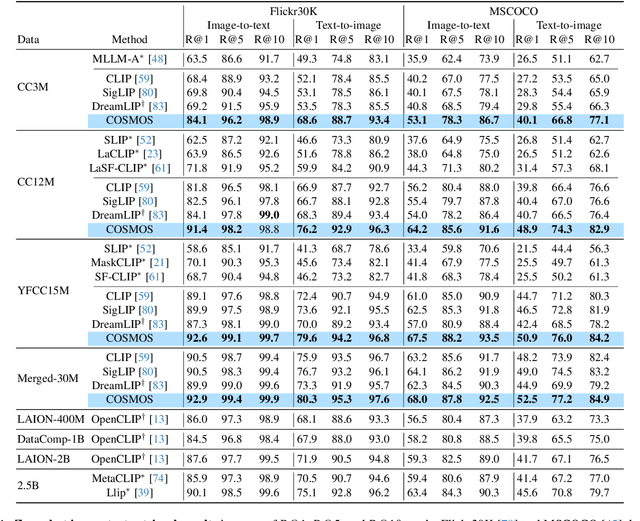
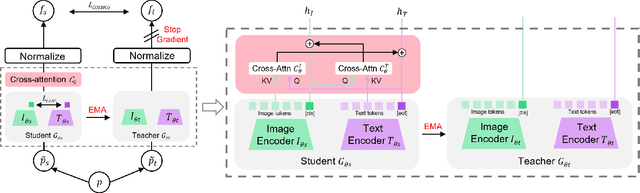
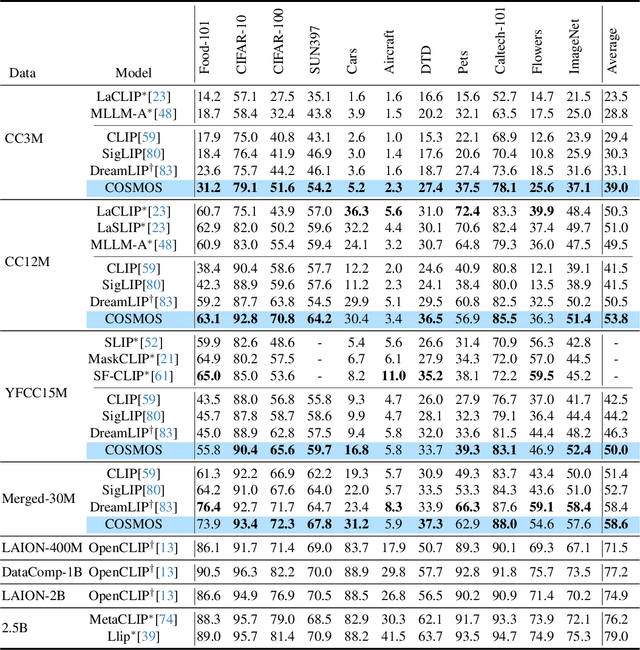
Abstract:Vision-Language Models (VLMs) trained with contrastive loss have achieved significant advancements in various vision and language tasks. However, the global nature of contrastive loss makes VLMs focus predominantly on foreground objects, neglecting other crucial information in the image, which limits their effectiveness in downstream tasks. To address these challenges, we propose COSMOS: CrOSs-MOdality Self-distillation for vision-language pre-training that integrates a novel text-cropping strategy and cross-attention module into a self-supervised learning framework. We create global and local views of images and texts (i.e., multi-modal augmentations), which are essential for self-distillation in VLMs. We further introduce a cross-attention module, enabling COSMOS to learn comprehensive cross-modal representations optimized via a cross-modality self-distillation loss. COSMOS consistently outperforms previous strong baselines on various zero-shot downstream tasks, including retrieval, classification, and semantic segmentation. Additionally, it surpasses CLIP-based models trained on larger datasets in visual perception and contextual understanding tasks.
Accurate Explanation Model for Image Classifiers using Class Association Embedding
Jun 12, 2024Abstract:Image classification is a primary task in data analysis where explainable models are crucially demanded in various applications. Although amounts of methods have been proposed to obtain explainable knowledge from the black-box classifiers, these approaches lack the efficiency of extracting global knowledge regarding the classification task, thus is vulnerable to local traps and often leads to poor accuracy. In this study, we propose a generative explanation model that combines the advantages of global and local knowledge for explaining image classifiers. We develop a representation learning method called class association embedding (CAE), which encodes each sample into a pair of separated class-associated and individual codes. Recombining the individual code of a given sample with altered class-associated code leads to a synthetic real-looking sample with preserved individual characters but modified class-associated features and possibly flipped class assignments. A building-block coherency feature extraction algorithm is proposed that efficiently separates class-associated features from individual ones. The extracted feature space forms a low-dimensional manifold that visualizes the classification decision patterns. Explanation on each individual sample can be then achieved in a counter-factual generation manner which continuously modifies the sample in one direction, by shifting its class-associated code along a guided path, until its classification outcome is changed. We compare our method with state-of-the-art ones on explaining image classification tasks in the form of saliency maps, demonstrating that our method achieves higher accuracies. The code is available at https://github.com/xrt11/XAI-CODE.
Active Globally Explainable Learning for Medical Images via Class Association Embedding and Cyclic Adversarial Generation
Jun 12, 2023Abstract:Explainability poses a major challenge to artificial intelligence (AI) techniques. Current studies on explainable AI (XAI) lack the efficiency of extracting global knowledge about the learning task, thus suffer deficiencies such as imprecise saliency, context-aware absence and vague meaning. In this paper, we propose the class association embedding (CAE) approach to address these issues. We employ an encoder-decoder architecture to embed sample features and separate them into class-related and individual-related style vectors simultaneously. Recombining the individual-style code of a given sample with the class-style code of another leads to a synthetic sample with preserved individual characters but changed class assignment, following a cyclic adversarial learning strategy. Class association embedding distills the global class-related features of all instances into a unified domain with well separation between classes. The transition rules between different classes can be then extracted and further employed to individual instances. We then propose an active XAI framework which manipulates the class-style vector of a certain sample along guided paths towards the counter-classes, resulting in a series of counter-example synthetic samples with identical individual characters. Comparing these counterfactual samples with the original ones provides a global, intuitive illustration to the nature of the classification tasks. We adopt the framework on medical image classification tasks, which show that more precise saliency maps with powerful context-aware representation can be achieved compared with existing methods. Moreover, the disease pathology can be directly visualized via traversing the paths in the class-style space.
SCOPE: Structural Continuity Preservation for Medical Image Segmentation
Apr 28, 2023Abstract:Although the preservation of shape continuity and physiological anatomy is a natural assumption in the segmentation of medical images, it is often neglected by deep learning methods that mostly aim for the statistical modeling of input data as pixels rather than interconnected structures. In biological structures, however, organs are not separate entities; for example, in reality, a severed vessel is an indication of an underlying problem, but traditional segmentation models are not designed to strictly enforce the continuity of anatomy, potentially leading to inaccurate medical diagnoses. To address this issue, we propose a graph-based approach that enforces the continuity and connectivity of anatomical topology in medical images. Our method encodes the continuity of shapes as a graph constraint, ensuring that the network's predictions maintain this continuity. We evaluate our method on two public benchmarks on retinal vessel segmentation, showing significant improvements in connectivity metrics compared to traditional methods while getting better or on-par performance on segmentation metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge