Yanqing Zhang
The Energy-Efficient Hierarchical Neural Network with Fast FPGA-Based Incremental Learning
Sep 18, 2025Abstract:The rising computational and energy demands of deep learning, particularly in large-scale architectures such as foundation models and large language models (LLMs), pose significant challenges to sustainability. Traditional gradient-based training methods are inefficient, requiring numerous iterative updates and high power consumption. To address these limitations, we propose a hybrid framework that combines hierarchical decomposition with FPGA-based direct equation solving and incremental learning. Our method divides the neural network into two functional tiers: lower layers are optimized via single-step equation solving on FPGAs for efficient and parallelizable feature extraction, while higher layers employ adaptive incremental learning to support continual updates without full retraining. Building upon this foundation, we introduce the Compound LLM framework, which explicitly deploys LLM modules across both hierarchy levels. The lower-level LLM handles reusable representation learning with minimal energy overhead, while the upper-level LLM performs adaptive decision-making through energy-aware updates. This integrated design enhances scalability, reduces redundant computation, and aligns with the principles of sustainable AI. Theoretical analysis and architectural insights demonstrate that our method reduces computational costs significantly while preserving high model performance, making it well-suited for edge deployment and real-time adaptation in energy-constrained environments.
Forecasting and Visualizing Air Quality from Sky Images with Vision-Language Models
Sep 18, 2025Abstract:Air pollution remains a critical threat to public health and environmental sustainability, yet conventional monitoring systems are often constrained by limited spatial coverage and accessibility. This paper proposes an AI-driven agent that predicts ambient air pollution levels from sky images and synthesizes realistic visualizations of pollution scenarios using generative modeling. Our approach combines statistical texture analysis with supervised learning for pollution classification, and leverages vision-language model (VLM)-guided image generation to produce interpretable representations of air quality conditions. The generated visuals simulate varying degrees of pollution, offering a foundation for user-facing interfaces that improve transparency and support informed environmental decision-making. These outputs can be seamlessly integrated into intelligent applications aimed at enhancing situational awareness and encouraging behavioral responses based on real-time forecasts. We validate our method using a dataset of urban sky images and demonstrate its effectiveness in both pollution level estimation and semantically consistent visual synthesis. The system design further incorporates human-centered user experience principles to ensure accessibility, clarity, and public engagement in air quality forecasting. To support scalable and energy-efficient deployment, future iterations will incorporate a green CNN architecture enhanced with FPGA-based incremental learning, enabling real-time inference on edge platforms.
BoolGebra: Attributed Graph-learning for Boolean Algebraic Manipulation
Jan 19, 2024



Abstract:Boolean algebraic manipulation is at the core of logic synthesis in Electronic Design Automation (EDA) design flow. Existing methods struggle to fully exploit optimization opportunities, and often suffer from an explosive search space and limited scalability efficiency. This work presents BoolGebra, a novel attributed graph-learning approach for Boolean algebraic manipulation that aims to improve fundamental logic synthesis. BoolGebra incorporates Graph Neural Networks (GNNs) and takes initial feature embeddings from both structural and functional information as inputs. A fully connected neural network is employed as the predictor for direct optimization result predictions, significantly reducing the search space and efficiently locating the optimization space. The experiments involve training the BoolGebra model w.r.t design-specific and cross-design inferences using the trained model, where BoolGebra demonstrates generalizability for cross-design inference and its potential to scale from small, simple training datasets to large, complex inference datasets. Finally, BoolGebra is integrated with existing synthesis tool ABC to perform end-to-end logic minimization evaluation w.r.t SOTA baselines.
GATSPI: GPU Accelerated Gate-Level Simulation for Power Improvement
Mar 11, 2022



Abstract:In this paper, we present GATSPI, a novel GPU accelerated logic gate simulator that enables ultra-fast power estimation for industry sized ASIC designs with millions of gates. GATSPI is written in PyTorch with custom CUDA kernels for ease of coding and maintainability. It achieves simulation kernel speedup of up to 1668X on a single-GPU system and up to 7412X on a multiple-GPU system when compared to a commercial gate-level simulator running on a single CPU core. GATSPI supports a range of simple to complex cell types from an industry standard cell library and SDF conditional delay statements without requiring prior calibration runs and produces industry-standard SAIF files from delay-aware gate-level simulation. Finally, we deploy GATSPI in a glitch-optimization flow, achieving a 1.4% power saving with a 449X speedup in turnaround time compared to a similar flow using a commercial simulator.
Covid-19 Detection from Chest X-ray and Patient Metadata using Graph Convolutional Neural Networks
May 21, 2021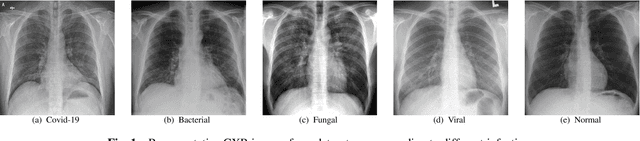
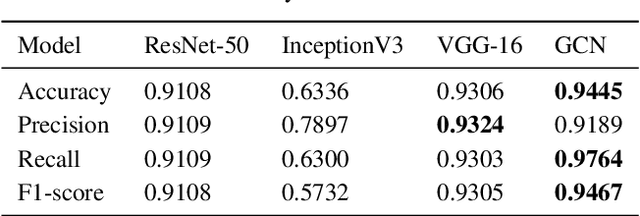
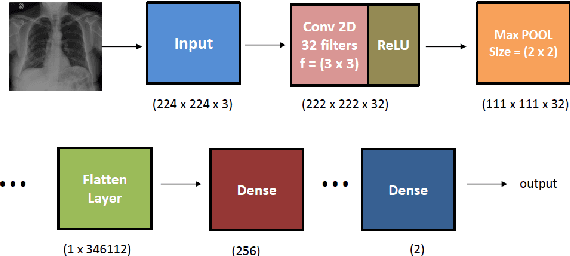
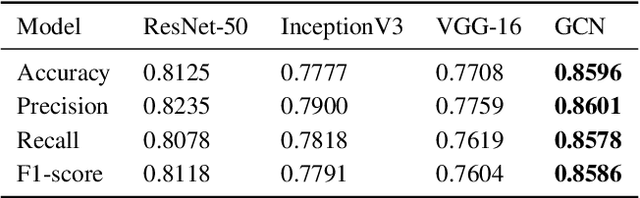
Abstract:The novel corona virus (Covid-19) has introduced significant challenges due to its rapid spreading nature through respiratory transmission. As a result, there is a huge demand for Artificial Intelligence (AI) based quick disease diagnosis methods as an alternative to high demand tests such as Polymerase Chain Reaction (PCR). Chest X-ray (CXR) Image analysis is such cost-effective radiography technique due to resource availability and quick screening. But, a sufficient and systematic data collection that is required by complex deep leaning (DL) models is more difficult and hence there are recent efforts that utilize transfer learning to address this issue. Still these transfer learnt models suffer from lack of generalization and increased bias to the training dataset resulting poor performance for unseen data. Limited correlation of the transferred features from the pre-trained model to a specific medical imaging domain like X-ray and overfitting on fewer data can be reasons for this circumstance. In this work, we propose a novel Graph Convolution Neural Network (GCN) that is capable of identifying bio-markers of Covid-19 pneumonia from CXR images and meta information about patients. The proposed method exploits important relational knowledge between data instances and their features using graph representation and applies convolution to learn the graph data which is not possible with conventional convolution on Euclidean domain. The results of extensive experiments of proposed model on binary (Covid vs normal) and three class (Covid, normal, other pneumonia) classification problems outperform different benchmark transfer learnt models, hence overcoming the aforementioned drawbacks.
FIST: A Feature-Importance Sampling and Tree-Based Method for Automatic Design Flow Parameter Tuning
Nov 26, 2020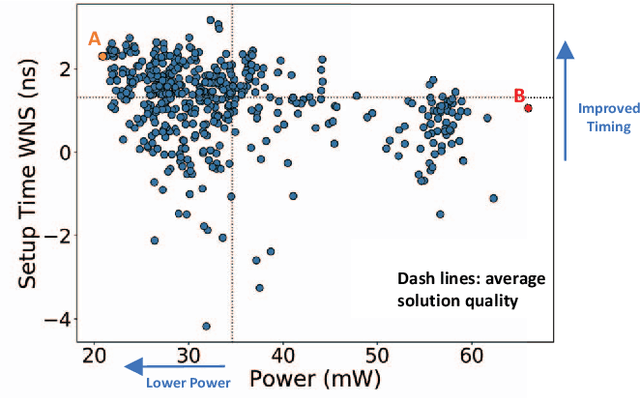
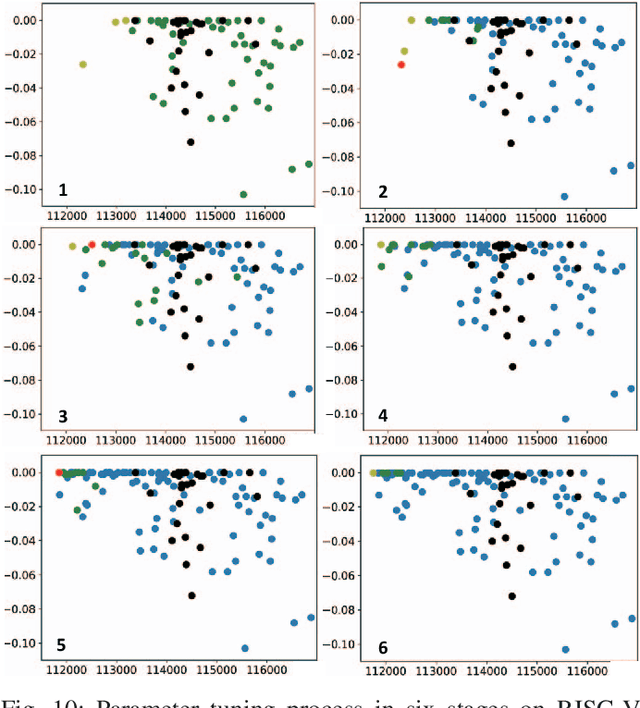
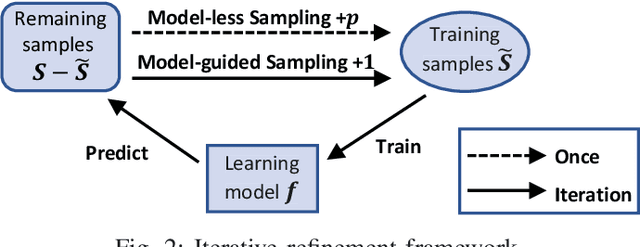
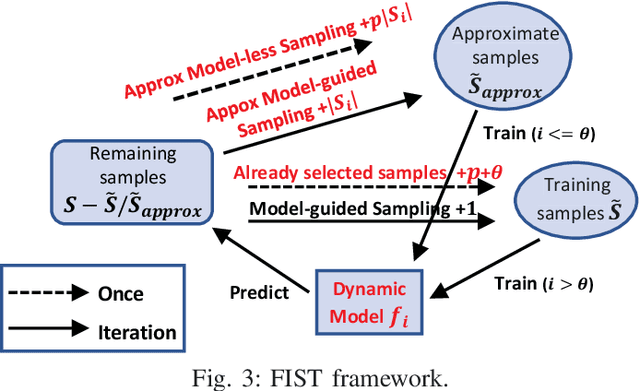
Abstract:Design flow parameters are of utmost importance to chip design quality and require a painfully long time to evaluate their effects. In reality, flow parameter tuning is usually performed manually based on designers' experience in an ad hoc manner. In this work, we introduce a machine learning-based automatic parameter tuning methodology that aims to find the best design quality with a limited number of trials. Instead of merely plugging in machine learning engines, we develop clustering and approximate sampling techniques for improving tuning efficiency. The feature extraction in this method can reuse knowledge from prior designs. Furthermore, we leverage a state-of-the-art XGBoost model and propose a novel dynamic tree technique to overcome overfitting. Experimental results on benchmark circuits show that our approach achieves 25% improvement in design quality or 37% reduction in sampling cost compared to random forest method, which is the kernel of a highly cited previous work. Our approach is further validated on two industrial designs. By sampling less than 0.02% of possible parameter sets, it reduces area by 1.83% and 1.43% compared to the best solutions hand-tuned by experienced designers.
Graph Convolution Networks Using Message Passing and Multi-Source Similarity Features for Predicting circRNA-Disease Association
Sep 15, 2020



Abstract:Graphs can be used to effectively represent complex data structures. Learning these irregular data in graphs is challenging and still suffers from shallow learning. Applying deep learning on graphs has recently showed good performance in many applications in social analysis, bioinformatics etc. A message passing graph convolution network is such a powerful method which has expressive power to learn graph structures. Meanwhile, circRNA is a type of non-coding RNA which plays a critical role in human diseases. Identifying the associations between circRNAs and diseases is important to diagnosis and treatment of complex diseases. However, there are limited number of known associations between them and conducting biological experiments to identify new associations is time consuming and expensive. As a result, there is a need of building efficient and feasible computation methods to predict potential circRNA-disease associations. In this paper, we propose a novel graph convolution network framework to learn features from a graph built with multi-source similarity information to predict circRNA-disease associations. First we use multi-source information of circRNA similarity, disease and circRNA Gaussian Interaction Profile (GIP) kernel similarity to extract the features using first graph convolution. Then we predict disease associations for each circRNA with second graph convolution. Proposed framework with five-fold cross validation on various experiments shows promising results in predicting circRNA-disease association and outperforms other existing methods.
Multicategory Angle-based Learning for Estimating Optimal Dynamic Treatment Regimes with Censored Data
Jan 14, 2020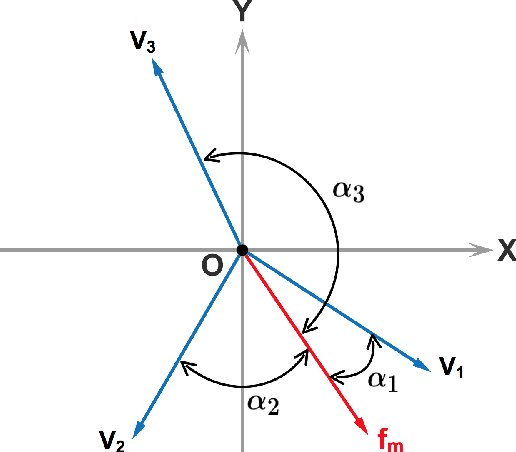
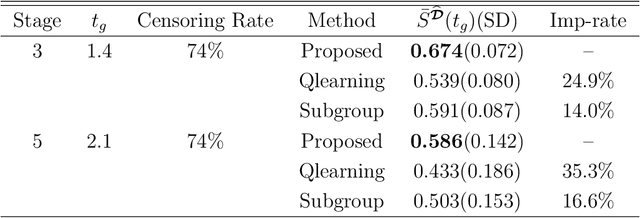
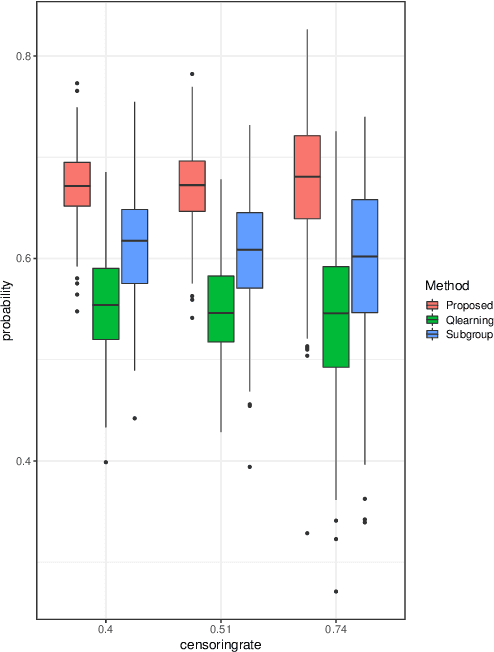

Abstract:An optimal dynamic treatment regime (DTR) consists of a sequence of decision rules in maximizing long-term benefits, which is applicable for chronic diseases such as HIV infection or cancer. In this paper, we develop a novel angle-based approach to search the optimal DTR under a multicategory treatment framework for survival data. The proposed method targets maximization the conditional survival function of patients following a DTR. In contrast to most existing approaches which are designed to maximize the expected survival time under a binary treatment framework, the proposed method solves the multicategory treatment problem given multiple stages for censored data. Specifically, the proposed method obtains the optimal DTR via integrating estimations of decision rules at multiple stages into a single multicategory classification algorithm without imposing additional constraints, which is also more computationally efficient and robust. In theory, we establish Fisher consistency of the proposed method under regularity conditions. Our numerical studies show that the proposed method outperforms competing methods in terms of maximizing the conditional survival function. We apply the proposed method to two real datasets: Framingham heart study data and acquired immunodeficiency syndrome (AIDS) clinical data.
Understanding 3D CNN Behavior for Alzheimer's Disease Diagnosis from Brain PET Scan
Dec 26, 2019


Abstract:In recent days, Convolutional Neural Networks (CNN) have demonstrated impressive performance in medical image analysis. However, there is a lack of clear understanding of why and how the Convolutional Neural Network performs so well for image analysis task. How CNN analyzes an image and discriminates among samples of different classes are usually considered as non-transparent. As a result, it becomes difficult to apply CNN based approaches in clinical procedures and automated disease diagnosis systems. In this paper, we consider this issue and work on visualizing and understanding the decision of Convolutional Neural Network for Alzheimer's Disease (AD) Diagnosis. We develop a 3D deep convolutional neural network for AD diagnosis using brain PET scans and propose using five visualizations techniques - Sensitivity Analysis (Backpropagation), Guided Backpropagation, Occlusion, Brain Area Occlusion, and Layer-wise Relevance Propagation (LRP) to understand the decision of the CNN by highlighting the relevant areas in the PET data.
Deep Learning Approach for Predicting 30 Day Readmissions after Coronary Artery Bypass Graft Surgery
Dec 03, 2018



Abstract:Hospital Readmissions within 30 days after discharge following Coronary Artery Bypass Graft (CABG) Surgery are substantial contributors to healthcare costs. Many predictive models were developed to identify risk factors for readmissions. However, majority of the existing models use statistical analysis techniques with data available at discharge. We propose an ensembled model to predict CABG readmissions using pre-discharge perioperative data and machine learning survival analysis techniques. Firstly, we applied fifty one potential readmission risk variables to Cox Proportional Hazard (CPH) survival regression univariate analysis. Fourteen of them turned out to be significant (with p value < 0.05), contributing to readmissions. Subsequently, we applied these 14 predictors to multivariate CPH model and Deep Learning Neural Network (NN) representation of the CPH model, DeepSurv. We validated this new ensembled model with 453 isolated adult CABG cases. Nine of the fourteen perioperative risk variables were identified as the most significant with Hazard Ratios (HR) of greater than 1.0. The concordance index metrics for CPH, DeepSurv, and ensembled models were then evaluated with training and validation datasets. Our ensembled model yielded promising results in terms of c-statistics, as we raised the the number of iterations and data set sizes. 30 day all-cause readmissions among isolated CABG patients can be predicted more effectively with perioperative pre-discharge data, using machine learning survival analysis techniques. Prediction accuracy levels could be improved further with deep learning algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge