Thosini Bamunu Mudiyanselage
Covid-19 Detection from Chest X-ray and Patient Metadata using Graph Convolutional Neural Networks
May 21, 2021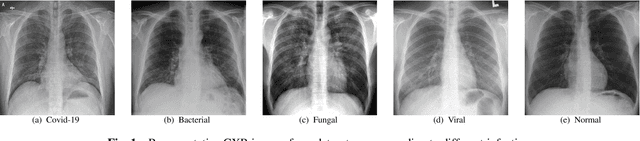
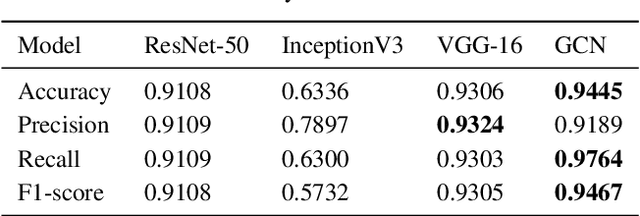
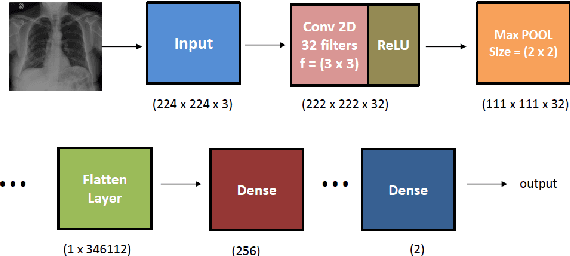
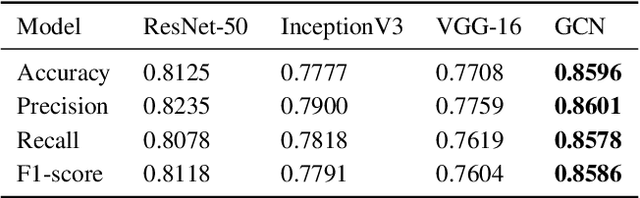
Abstract:The novel corona virus (Covid-19) has introduced significant challenges due to its rapid spreading nature through respiratory transmission. As a result, there is a huge demand for Artificial Intelligence (AI) based quick disease diagnosis methods as an alternative to high demand tests such as Polymerase Chain Reaction (PCR). Chest X-ray (CXR) Image analysis is such cost-effective radiography technique due to resource availability and quick screening. But, a sufficient and systematic data collection that is required by complex deep leaning (DL) models is more difficult and hence there are recent efforts that utilize transfer learning to address this issue. Still these transfer learnt models suffer from lack of generalization and increased bias to the training dataset resulting poor performance for unseen data. Limited correlation of the transferred features from the pre-trained model to a specific medical imaging domain like X-ray and overfitting on fewer data can be reasons for this circumstance. In this work, we propose a novel Graph Convolution Neural Network (GCN) that is capable of identifying bio-markers of Covid-19 pneumonia from CXR images and meta information about patients. The proposed method exploits important relational knowledge between data instances and their features using graph representation and applies convolution to learn the graph data which is not possible with conventional convolution on Euclidean domain. The results of extensive experiments of proposed model on binary (Covid vs normal) and three class (Covid, normal, other pneumonia) classification problems outperform different benchmark transfer learnt models, hence overcoming the aforementioned drawbacks.
PK-GCN: Prior Knowledge Assisted Image Classification using Graph Convolution Networks
Sep 24, 2020
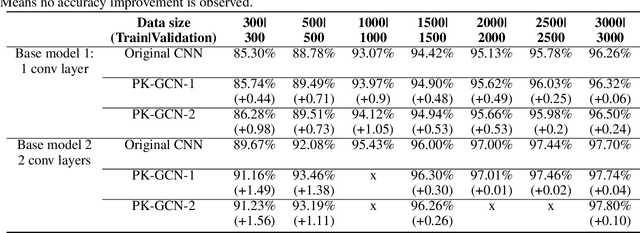
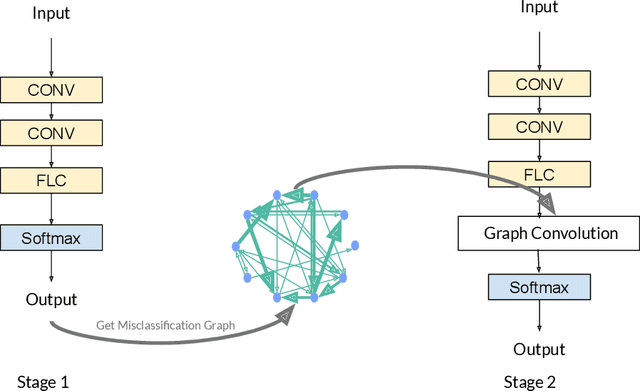
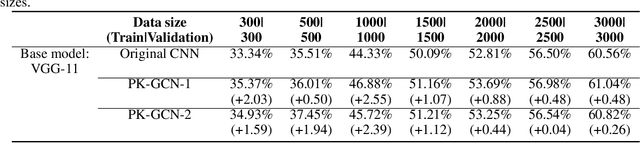
Abstract:Deep learning has gained great success in various classification tasks. Typically, deep learning models learn underlying features directly from data, and no underlying relationship between classes are included. Similarity between classes can influence the performance of classification. In this article, we propose a method that incorporates class similarity knowledge into convolutional neural networks models using a graph convolution layer. We evaluate our method on two benchmark image datasets: MNIST and CIFAR10, and analyze the results on different data and model sizes. Experimental results show that our model can improve classification accuracy, especially when the amount of available data is small.
Graph Convolution Networks Using Message Passing and Multi-Source Similarity Features for Predicting circRNA-Disease Association
Sep 15, 2020



Abstract:Graphs can be used to effectively represent complex data structures. Learning these irregular data in graphs is challenging and still suffers from shallow learning. Applying deep learning on graphs has recently showed good performance in many applications in social analysis, bioinformatics etc. A message passing graph convolution network is such a powerful method which has expressive power to learn graph structures. Meanwhile, circRNA is a type of non-coding RNA which plays a critical role in human diseases. Identifying the associations between circRNAs and diseases is important to diagnosis and treatment of complex diseases. However, there are limited number of known associations between them and conducting biological experiments to identify new associations is time consuming and expensive. As a result, there is a need of building efficient and feasible computation methods to predict potential circRNA-disease associations. In this paper, we propose a novel graph convolution network framework to learn features from a graph built with multi-source similarity information to predict circRNA-disease associations. First we use multi-source information of circRNA similarity, disease and circRNA Gaussian Interaction Profile (GIP) kernel similarity to extract the features using first graph convolution. Then we predict disease associations for each circRNA with second graph convolution. Proposed framework with five-fold cross validation on various experiments shows promising results in predicting circRNA-disease association and outperforms other existing methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge