Yangqin Feng
Are Traditional Deep Learning Model Approaches as Effective as a Retinal-Specific Foundation Model for Ocular and Systemic Disease Detection?
Jan 21, 2025Abstract:Background: RETFound, a self-supervised, retina-specific foundation model (FM), showed potential in downstream applications. However, its comparative performance with traditional deep learning (DL) models remains incompletely understood. This study aimed to evaluate RETFound against three ImageNet-pretrained supervised DL models (ResNet50, ViT-base, SwinV2) in detecting ocular and systemic diseases. Methods: We fine-tuned/trained RETFound and three DL models on full datasets, 50%, 20%, and fixed sample sizes (400, 200, 100 images, with half comprising disease cases; for each DR severity class, 100 and 50 cases were used. Fine-tuned models were tested internally using the SEED (53,090 images) and APTOS-2019 (3,672 images) datasets and externally validated on population-based (BES, CIEMS, SP2, UKBB) and open-source datasets (ODIR-5k, PAPILA, GAMMA, IDRiD, MESSIDOR-2). Model performance was compared using area under the receiver operating characteristic curve (AUC) and Z-tests with Bonferroni correction (P<0.05/3). Interpretation: Traditional DL models are mostly comparable to RETFound for ocular disease detection with large datasets. However, RETFound is superior in systemic disease detection with smaller datasets. These findings offer valuable insights into the respective merits and limitation of traditional models and FMs.
Enhancing Community Vision Screening -- AI Driven Retinal Photography for Early Disease Detection and Patient Trust
Oct 27, 2024Abstract:Community vision screening plays a crucial role in identifying individuals with vision loss and preventing avoidable blindness, particularly in rural communities where access to eye care services is limited. Currently, there is a pressing need for a simple and efficient process to screen and refer individuals with significant eye disease-related vision loss to tertiary eye care centers for further care. An ideal solution should seamlessly and readily integrate with existing workflows, providing comprehensive initial screening results to service providers, thereby enabling precise patient referrals for timely treatment. This paper introduces the Enhancing Community Vision Screening (ECVS) solution, which addresses the aforementioned concerns with a novel and feasible solution based on simple, non-invasive retinal photography for the detection of pathology-based visual impairment. Our study employs four distinct deep learning models: RETinal photo Quality Assessment (RETQA), Pathology Visual Impairment detection (PVI), Eye Disease Diagnosis (EDD) and Visualization of Lesion Regions of the eye (VLR). We conducted experiments on over 10 datasets, totaling more than 80,000 fundus photos collected from various sources. The models integrated into ECVS achieved impressive AUC scores of 0.98 for RETQA, 0.95 for PVI, and 0.90 for EDD, along with a DICE coefficient of 0.48 for VLR. These results underscore the promising capabilities of ECVS as a straightforward and scalable method for community-based vision screening.
Localizing Anatomical Landmarks in Ocular Images using Zoom-In Attentive Networks
Sep 25, 2022Abstract:Localizing anatomical landmarks are important tasks in medical image analysis. However, the landmarks to be localized often lack prominent visual features. Their locations are elusive and easily confused with the background, and thus precise localization highly depends on the context formed by their surrounding areas. In addition, the required precision is usually higher than segmentation and object detection tasks. Therefore, localization has its unique challenges different from segmentation or detection. In this paper, we propose a zoom-in attentive network (ZIAN) for anatomical landmark localization in ocular images. First, a coarse-to-fine, or "zoom-in" strategy is utilized to learn the contextualized features in different scales. Then, an attentive fusion module is adopted to aggregate multi-scale features, which consists of 1) a co-attention network with a multiple regions-of-interest (ROIs) scheme that learns complementary features from the multiple ROIs, 2) an attention-based fusion module which integrates the multi-ROIs features and non-ROI features. We evaluated ZIAN on two open challenge tasks, i.e., the fovea localization in fundus images and scleral spur localization in AS-OCT images. Experiments show that ZIAN achieves promising performances and outperforms state-of-the-art localization methods. The source code and trained models of ZIAN are available at https://github.com/leixiaofeng-astar/OMIA9-ZIAN.
Few-Shot Domain Adaptation with Polymorphic Transformers
Jul 10, 2021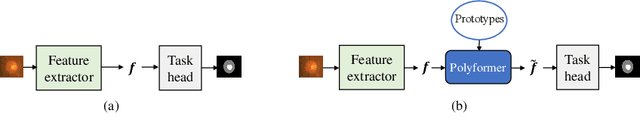
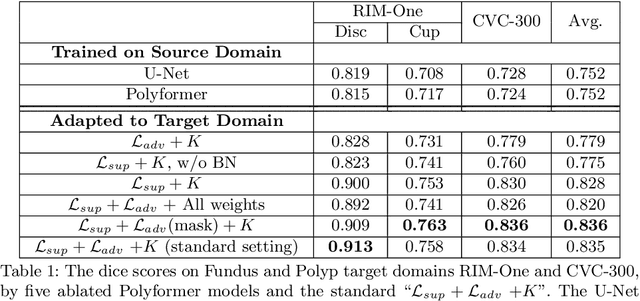
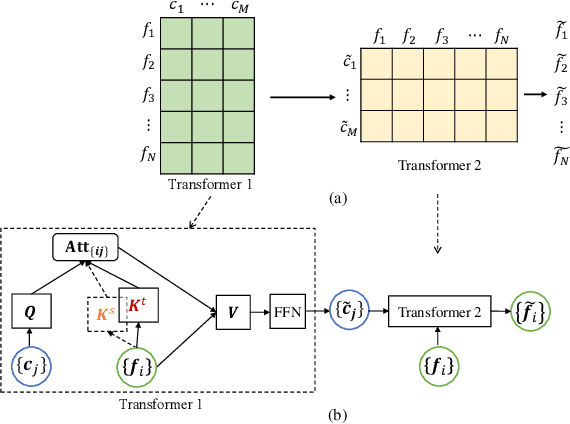
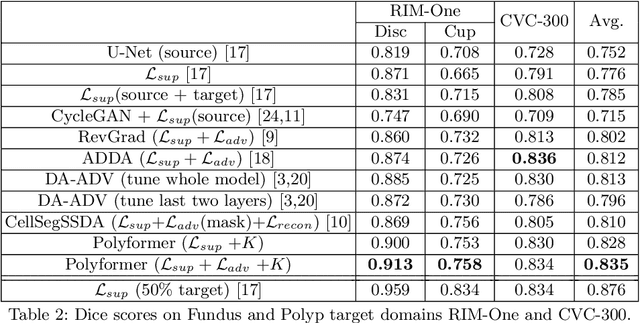
Abstract:Deep neural networks (DNNs) trained on one set of medical images often experience severe performance drop on unseen test images, due to various domain discrepancy between the training images (source domain) and the test images (target domain), which raises a domain adaptation issue. In clinical settings, it is difficult to collect enough annotated target domain data in a short period. Few-shot domain adaptation, i.e., adapting a trained model with a handful of annotations, is highly practical and useful in this case. In this paper, we propose a Polymorphic Transformer (Polyformer), which can be incorporated into any DNN backbones for few-shot domain adaptation. Specifically, after the polyformer layer is inserted into a model trained on the source domain, it extracts a set of prototype embeddings, which can be viewed as a "basis" of the source-domain features. On the target domain, the polyformer layer adapts by only updating a projection layer which controls the interactions between image features and the prototype embeddings. All other model weights (except BatchNorm parameters) are frozen during adaptation. Thus, the chance of overfitting the annotations is greatly reduced, and the model can perform robustly on the target domain after being trained on a few annotated images. We demonstrate the effectiveness of Polyformer on two medical segmentation tasks (i.e., optic disc/cup segmentation, and polyp segmentation). The source code of Polyformer is released at https://github.com/askerlee/segtran.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge