Mingrui Tan
Note2Chat: Improving LLMs for Multi-Turn Clinical History Taking Using Medical Notes
Jan 29, 2026Abstract:Effective clinical history taking is a foundational yet underexplored component of clinical reasoning. While large language models (LLMs) have shown promise on static benchmarks, they often fall short in dynamic, multi-turn diagnostic settings that require iterative questioning and hypothesis refinement. To address this gap, we propose \method{}, a note-driven framework that trains LLMs to conduct structured history taking and diagnosis by learning from widely available medical notes. Instead of relying on scarce and sensitive dialogue data, we convert real-world medical notes into high-quality doctor-patient dialogues using a decision tree-guided generation and refinement pipeline. We then propose a three-stage fine-tuning strategy combining supervised learning, simulated data augmentation, and preference learning. Furthermore, we propose a novel single-turn reasoning paradigm that reframes history taking as a sequence of single-turn reasoning problems. This design enhances interpretability and enables local supervision, dynamic adaptation, and greater sample efficiency. Experimental results show that our method substantially improves clinical reasoning, achieving gains of +16.9 F1 and +21.0 Top-1 diagnostic accuracy over GPT-4o. Our code and dataset can be found at https://github.com/zhentingsheng/Note2Chat.
Confidence-Based Task Prediction in Continual Disease Classification Using Probability Distribution
Jun 03, 2024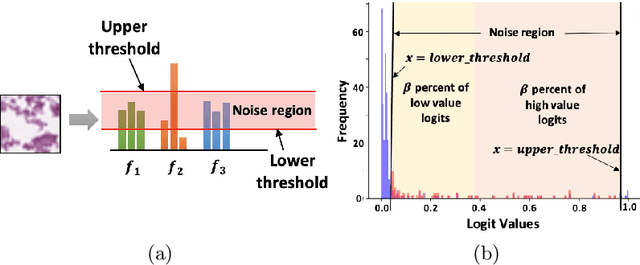
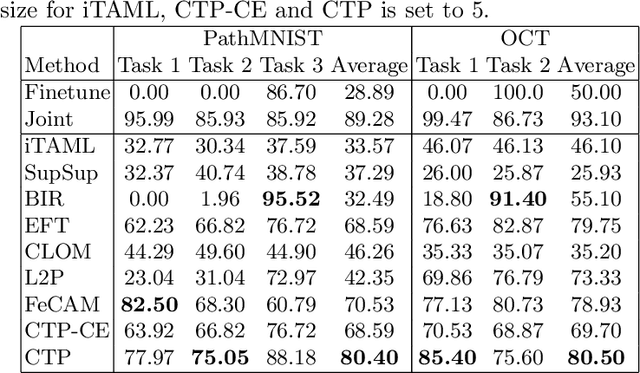
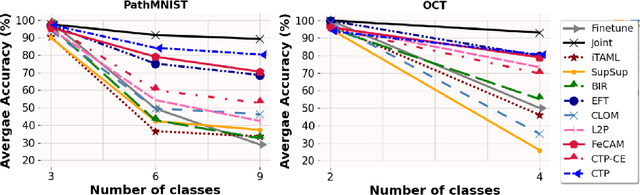
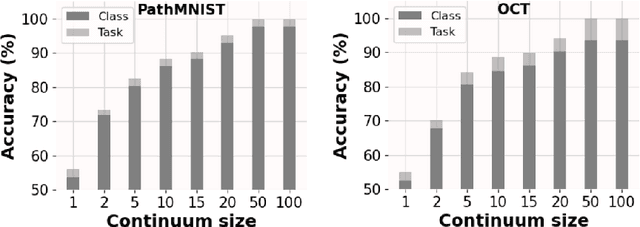
Abstract:Deep learning models are widely recognized for their effectiveness in identifying medical image findings in disease classification. However, their limitations become apparent in the dynamic and ever-changing clinical environment, characterized by the continuous influx of newly annotated medical data from diverse sources. In this context, the need for continual learning becomes particularly paramount, not only to adapt to evolving medical scenarios but also to ensure the privacy of healthcare data. In our research, we emphasize the utilization of a network comprising expert classifiers, where a new expert classifier is added each time a new task is introduced. We present CTP, a task-id predictor that utilizes confidence scores, leveraging the probability distribution (logits) of the classifier to accurately determine the task-id at inference time. Logits are adjusted to ensure that classifiers yield a high-entropy distribution for data associated with tasks other than their own. By defining a noise region in the distribution and computing confidence scores, CTP achieves superior performance when compared to other relevant continual learning methods. Additionally, the performance of CTP can be further improved by providing it with a continuum of data at the time of inference.
Localizing Anatomical Landmarks in Ocular Images using Zoom-In Attentive Networks
Sep 25, 2022Abstract:Localizing anatomical landmarks are important tasks in medical image analysis. However, the landmarks to be localized often lack prominent visual features. Their locations are elusive and easily confused with the background, and thus precise localization highly depends on the context formed by their surrounding areas. In addition, the required precision is usually higher than segmentation and object detection tasks. Therefore, localization has its unique challenges different from segmentation or detection. In this paper, we propose a zoom-in attentive network (ZIAN) for anatomical landmark localization in ocular images. First, a coarse-to-fine, or "zoom-in" strategy is utilized to learn the contextualized features in different scales. Then, an attentive fusion module is adopted to aggregate multi-scale features, which consists of 1) a co-attention network with a multiple regions-of-interest (ROIs) scheme that learns complementary features from the multiple ROIs, 2) an attention-based fusion module which integrates the multi-ROIs features and non-ROI features. We evaluated ZIAN on two open challenge tasks, i.e., the fovea localization in fundus images and scleral spur localization in AS-OCT images. Experiments show that ZIAN achieves promising performances and outperforms state-of-the-art localization methods. The source code and trained models of ZIAN are available at https://github.com/leixiaofeng-astar/OMIA9-ZIAN.
Benchmarking Predictive Risk Models for Emergency Departments with Large Public Electronic Health Records
Nov 22, 2021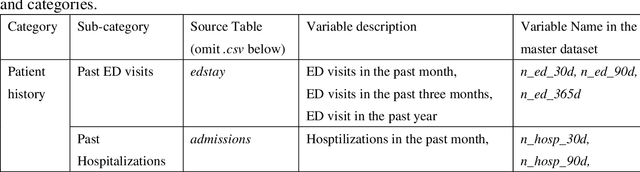
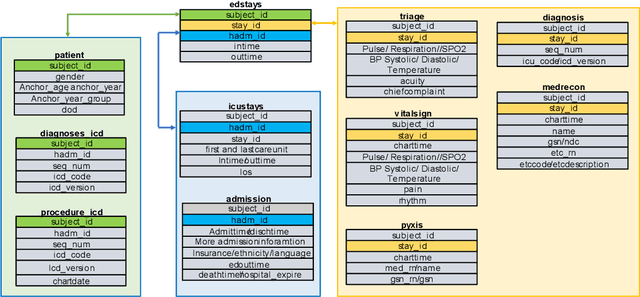
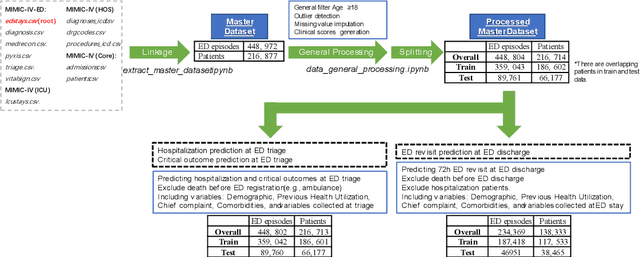
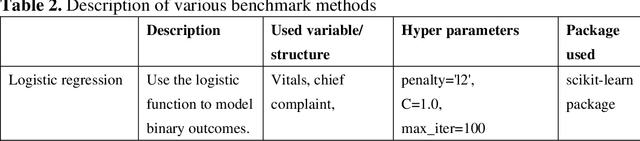
Abstract:There is a continuously growing demand for emergency department (ED) services across the world, especially under the COVID-19 pandemic. Risk triaging plays a crucial role in prioritizing limited medical resources for patients who need them most. Recently the pervasive use of Electronic Health Records (EHR) has generated a large volume of stored data, accompanied by vast opportunities for the development of predictive models which could improve emergency care. However, there is an absence of widely accepted ED benchmarks based on large-scale public EHR, which new researchers could easily access. Success in filling in this gap could enable researchers to start studies on ED more quickly and conveniently without verbose data preprocessing and facilitate comparisons among different studies and methodologies. In this paper, based on the Medical Information Mart for Intensive Care IV Emergency Department (MIMIC-IV-ED) database, we proposed a public ED benchmark suite and obtained a benchmark dataset containing over 500,000 ED visits episodes from 2011 to 2019. Three ED-based prediction tasks (hospitalization, critical outcomes, and 72-hour ED revisit) were introduced, where various popular methodologies, from machine learning methods to clinical scoring systems, were implemented. The results of their performance were evaluated and compared. Our codes are open-source so that anyone with access to MIMIC-IV-ED could follow the same steps of data processing, build the benchmarks, and reproduce the experiments. This study provided insights, suggestions, as well as protocols for future researchers to process the raw data and quickly build up models for emergency care.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge