Xintian Han
LEMUR: Large scale End-to-end MUltimodal Recommendation
Nov 17, 2025Abstract:Traditional ID-based recommender systems often struggle with cold-start and generalization challenges. Multimodal recommendation systems, which leverage textual and visual data, offer a promising solution to mitigate these issues. However, existing industrial approaches typically adopt a two-stage training paradigm: first pretraining a multimodal model, then applying its frozen representations to train the recommendation model. This decoupled framework suffers from misalignment between multimodal learning and recommendation objectives, as well as an inability to adapt dynamically to new data. To address these limitations, we propose LEMUR, the first large-scale multimodal recommender system trained end-to-end from raw data. By jointly optimizing both the multimodal and recommendation components, LEMUR ensures tighter alignment with downstream objectives while enabling real-time parameter updates. Constructing multimodal sequential representations from user history often entails prohibitively high computational costs. To alleviate this bottleneck, we propose a novel memory bank mechanism that incrementally accumulates historical multimodal representations throughout the training process. After one month of deployment in Douyin Search, LEMUR has led to a 0.843% reduction in query change rate decay and a 0.81% improvement in QAUC. Additionally, LEMUR has shown significant gains across key offline metrics for Douyin Advertisement. Our results validate the superiority of end-to-end multimodal recommendation in real-world industrial scenarios.
Real-time Indexing for Large-scale Recommendation by Streaming Vector Quantization Retriever
Jan 15, 2025Abstract:Retrievers, which form one of the most important recommendation stages, are responsible for efficiently selecting possible positive samples to the later stages under strict latency limitations. Because of this, large-scale systems always rely on approximate calculations and indexes to roughly shrink candidate scale, with a simple ranking model. Considering simple models lack the ability to produce precise predictions, most of the existing methods mainly focus on incorporating complicated ranking models. However, another fundamental problem of index effectiveness remains unresolved, which also bottlenecks complication. In this paper, we propose a novel index structure: streaming Vector Quantization model, as a new generation of retrieval paradigm. Streaming VQ attaches items with indexes in real time, granting it immediacy. Moreover, through meticulous verification of possible variants, it achieves additional benefits like index balancing and reparability, enabling it to support complicated ranking models as existing approaches. As a lightweight and implementation-friendly architecture, streaming VQ has been deployed and replaced all major retrievers in Douyin and Douyin Lite, resulting in remarkable user engagement gain.
Unlock the Correlation between Supervised Fine-Tuning and Reinforcement Learning in Training Code Large Language Models
Jun 14, 2024



Abstract:Automatic code generation has been a longstanding research topic. With the advancement of general-purpose large language models (LLMs), the ability to code stands out as one important measure to the model's reasoning performance. Usually, a two-stage training paradigm is implemented to obtain a Code LLM, namely the pretraining and the fine-tuning. Within the fine-tuning, supervised fine-tuning (SFT), and reinforcement learning (RL) are often used to improve the model's zero-shot ability. A large number of work has been conducted to improve the model's performance on code-related benchmarks with either modifications to the algorithm or refinement of the dataset. However, we still lack a deep insight into the correlation between SFT and RL. For instance, what kind of dataset should be used to ensure generalization, or what if we abandon the SFT phase in fine-tuning. In this work, we make an attempt to understand the correlation between SFT and RL. To facilitate our research, we manually craft 100 basis python functions, called atomic functions, and then a synthesizing pipeline is deployed to create a large number of synthetic functions on top of the atomic ones. In this manner, we ensure that the train and test sets remain distinct, preventing data contamination. Through comprehensive ablation study, we find: (1) Both atomic and synthetic functions are indispensable for SFT's generalization, and only a handful of synthetic functions are adequate; (2) Through RL, the SFT's generalization to target domain can be greatly enhanced, even with the same training prompts; (3) Training RL from scratch can alleviate the over-fitting issue introduced in the SFT phase.
Survival Mixture Density Networks
Aug 23, 2022
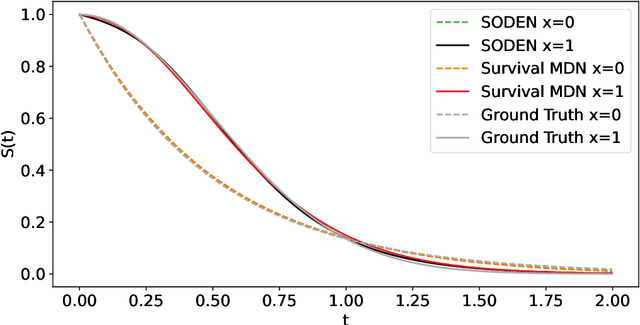

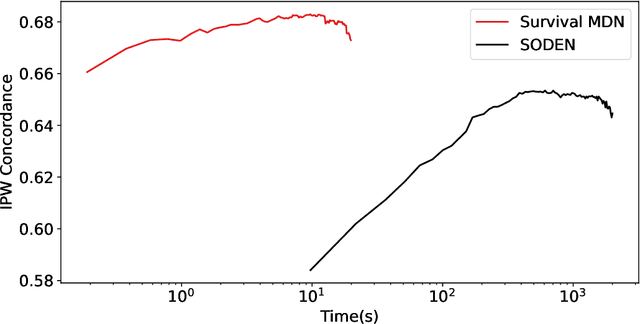
Abstract:Survival analysis, the art of time-to-event modeling, plays an important role in clinical treatment decisions. Recently, continuous time models built from neural ODEs have been proposed for survival analysis. However, the training of neural ODEs is slow due to the high computational complexity of neural ODE solvers. Here, we propose an efficient alternative for flexible continuous time models, called Survival Mixture Density Networks (Survival MDNs). Survival MDN applies an invertible positive function to the output of Mixture Density Networks (MDNs). While MDNs produce flexible real-valued distributions, the invertible positive function maps the model into the time-domain while preserving a tractable density. Using four datasets, we show that Survival MDN performs better than, or similarly to continuous and discrete time baselines on concordance, integrated Brier score and integrated binomial log-likelihood. Meanwhile, Survival MDNs are also faster than ODE-based models and circumvent binning issues in discrete models.
Inverse-Weighted Survival Games
Nov 16, 2021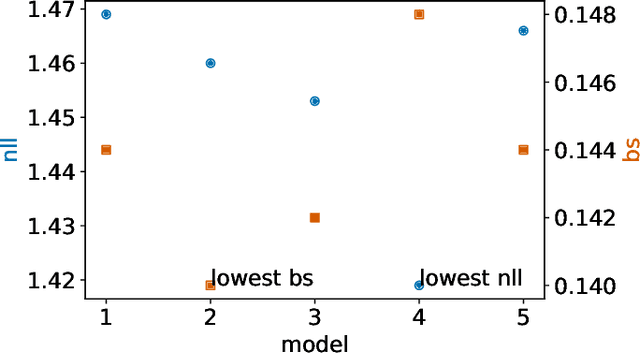
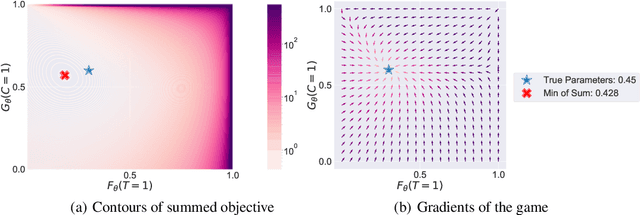
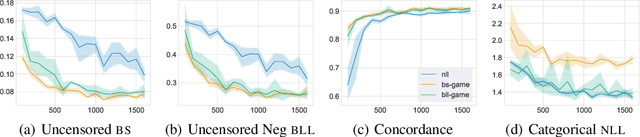
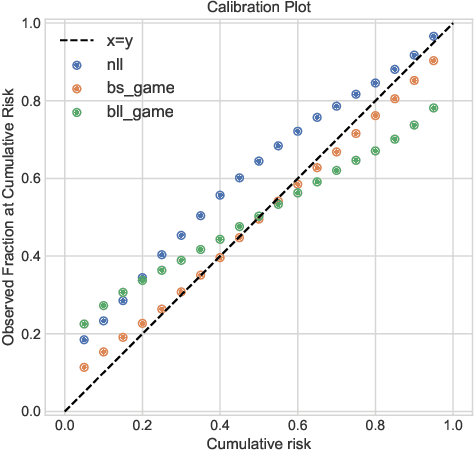
Abstract:Deep models trained through maximum likelihood have achieved state-of-the-art results for survival analysis. Despite this training scheme, practitioners evaluate models under other criteria, such as binary classification losses at a chosen set of time horizons, e.g. Brier score (BS) and Bernoulli log likelihood (BLL). Models trained with maximum likelihood may have poor BS or BLL since maximum likelihood does not directly optimize these criteria. Directly optimizing criteria like BS requires inverse-weighting by the censoring distribution, estimation of which itself also requires inverse-weighted by the failure distribution. But neither are known. To resolve this dilemma, we introduce Inverse-Weighted Survival Games to train both failure and censoring models with respect to criteria such as BS or BLL. In these games, objectives for each model are built from re-weighted estimates featuring the other model, where the re-weighting model is held fixed during training. When the loss is proper, we show that the games always have the true failure and censoring distributions as a stationary point. This means models in the game do not leave the correct distributions once reached. We construct one case where this stationary point is unique. We show that these games optimize BS on simulations and then apply these principles on real world cancer and critically-ill patient data.
X-CAL: Explicit Calibration for Survival Analysis
Jan 13, 2021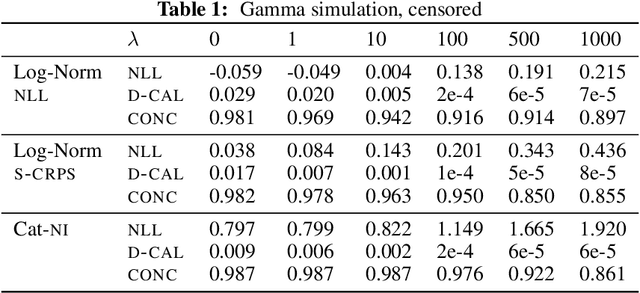
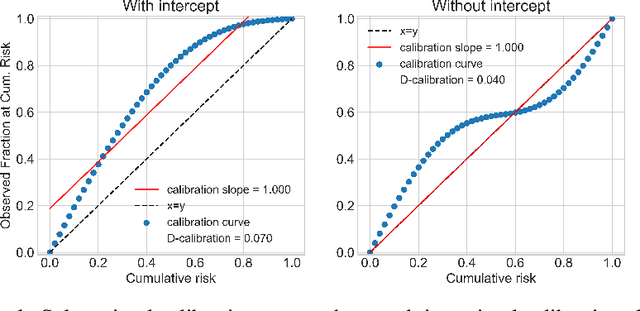
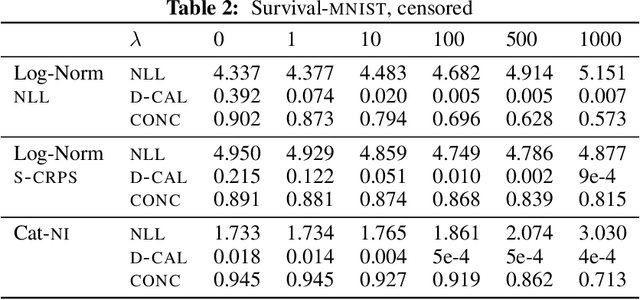
Abstract:Survival analysis models the distribution of time until an event of interest, such as discharge from the hospital or admission to the ICU. When a model's predicted number of events within any time interval is similar to the observed number, it is called well-calibrated. A survival model's calibration can be measured using, for instance, distributional calibration (D-CALIBRATION) [Haider et al., 2020] which computes the squared difference between the observed and predicted number of events within different time intervals. Classically, calibration is addressed in post-training analysis. We develop explicit calibration (X-CAL), which turns D-CALIBRATION into a differentiable objective that can be used in survival modeling alongside maximum likelihood estimation and other objectives. X-CAL allows practitioners to directly optimize calibration and strike a desired balance between predictive power and calibration. In our experiments, we fit a variety of shallow and deep models on simulated data, a survival dataset based on MNIST, on length-of-stay prediction using MIMIC-III data, and on brain cancer data from The Cancer Genome Atlas. We show that the models we study can be miscalibrated. We give experimental evidence on these datasets that X-CAL improves D-CALIBRATION without a large decrease in concordance or likelihood.
Explaining Data-Driven Decisions made by AI Systems: The Counterfactual Approach
Feb 05, 2020



Abstract:Lack of understanding of the decisions made by model-based AI systems is an important barrier for their adoption. We examine counterfactual explanations as an alternative for explaining AI decisions. The counterfactual approach defines an explanation as a set of the system's data inputs that causally drives the decision (meaning that removing them changes the decision) and is irreducible (meaning that removing any subset of the inputs in the explanation does not change the decision). We generalize previous work on counterfactual explanations, resulting in a framework that (a) is model-agnostic, (b) can address features with arbitrary data types, (c) can explain decisions made by complex AI systems that incorporate multiple models, and (d) is scalable to large numbers of features. We also propose a heuristic procedure to find the most useful explanations depending on the context. We contrast counterfactual explanations with another alternative: methods that explain model predictions by weighting features according to their importance (e.g., SHAP, LIME). This paper presents two fundamental reasons why explaining model predictions is not the same as explaining the decisions made using those predictions, suggesting we should carefully consider whether importance-weight explanations are well-suited to explain decisions made by AI systems. Specifically, we show that (1) features that have a large importance weight for a model prediction may not actually affect the corresponding decision, and (2) importance weights are insufficient to communicate whether and how features influence system decisions. We demonstrate this with several examples, including three detailed case studies that compare the counterfactual approach with SHAP to illustrate various conditions under which counterfactual explanations explain data-driven decisions better than feature importance weights.
Adversarial Examples for Electrocardiograms
Jun 04, 2019



Abstract:In recent years, the electrocardiogram (ECG) has seen a large diffusion in both medical and commercial applications, fueled by the rise of single-lead versions. Single-lead ECG can be embedded in medical devices and wearable products such as the injectable Medtronic Linq monitor, the iRhythm Ziopatch wearable monitor, and the Apple Watch Series 4. Recently, deep neural networks have been used to automatically analyze ECG tracings, outperforming even physicians specialized in cardiac electrophysiology in detecting certain rhythm irregularities. However, deep learning classifiers have been shown to be brittle to adversarial examples, which are examples created to look incontrovertibly belonging to a certain class to a human eye but contain subtle features that fool the classifier into misclassifying them into the wrong class. Very recently, adversarial examples have also been created for medical-related tasks. Yet, traditional attack methods to create adversarial examples, such as projected gradient descent (PGD) do not extend directly to ECG signals, as they generate examples that introduce square wave artifacts that are not physiologically plausible. Here, we developed a method to construct smoothed adversarial examples for single-lead ECG. First, we implemented a neural network model achieving state-of-the-art performance on the data from the 2017 PhysioNet/Computing-in-Cardiology Challenge for arrhythmia detection from single lead ECG classification. For this model, we utilized a new technique to generate smoothed examples to produce signals that are 1) indistinguishable to cardiologists from the original examples and 2) incorrectly classified by the neural network. Finally, we show that adversarial examples are not unique and provide a general technique to collate and perturb known adversarial examples to create new ones.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge