Xiaozhuang Song
MedGNN: Towards Multi-resolution Spatiotemporal Graph Learning for Medical Time Series Classification
Feb 06, 2025



Abstract:Medical time series has been playing a vital role in real-world healthcare systems as valuable information in monitoring health conditions of patients. Accurate classification for medical time series, e.g., Electrocardiography (ECG) signals, can help for early detection and diagnosis. Traditional methods towards medical time series classification rely on handcrafted feature extraction and statistical methods; with the recent advancement of artificial intelligence, the machine learning and deep learning methods have become more popular. However, existing methods often fail to fully model the complex spatial dynamics under different scales, which ignore the dynamic multi-resolution spatial and temporal joint inter-dependencies. Moreover, they are less likely to consider the special baseline wander problem as well as the multi-view characteristics of medical time series, which largely hinders their prediction performance. To address these limitations, we propose a Multi-resolution Spatiotemporal Graph Learning framework, MedGNN, for medical time series classification. Specifically, we first propose to construct multi-resolution adaptive graph structures to learn dynamic multi-scale embeddings. Then, to address the baseline wander problem, we propose Difference Attention Networks to operate self-attention mechanisms on the finite difference for temporal modeling. Moreover, to learn the multi-view characteristics, we utilize the Frequency Convolution Networks to capture complementary information of medical time series from the frequency domain. In addition, we introduce the Multi-resolution Graph Transformer architecture to model the dynamic dependencies and fuse the information from different resolutions. Finally, we have conducted extensive experiments on multiple medical real-world datasets that demonstrate the superior performance of our method. Our Code is available.
PTaRL: Prototype-based Tabular Representation Learning via Space Calibration
Jul 07, 2024
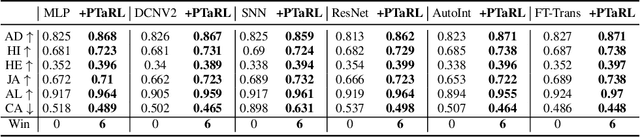

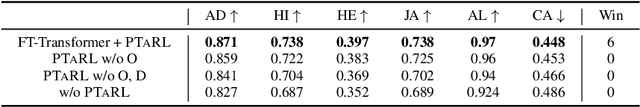
Abstract:Tabular data have been playing a mostly important role in diverse real-world fields, such as healthcare, engineering, finance, etc. With the recent success of deep learning, many tabular machine learning (ML) methods based on deep networks (e.g., Transformer, ResNet) have achieved competitive performance on tabular benchmarks. However, existing deep tabular ML methods suffer from the representation entanglement and localization, which largely hinders their prediction performance and leads to performance inconsistency on tabular tasks. To overcome these problems, we explore a novel direction of applying prototype learning for tabular ML and propose a prototype-based tabular representation learning framework, PTaRL, for tabular prediction tasks. The core idea of PTaRL is to construct prototype-based projection space (P-Space) and learn the disentangled representation around global data prototypes. Specifically, PTaRL mainly involves two stages: (i) Prototype Generation, that constructs global prototypes as the basis vectors of P-Space for representation, and (ii) Prototype Projection, that projects the data samples into P-Space and keeps the core global data information via Optimal Transport. Then, to further acquire the disentangled representations, we constrain PTaRL with two strategies: (i) to diversify the coordinates towards global prototypes of different representations within P-Space, we bring up a diversification constraint for representation calibration; (ii) to avoid prototype entanglement in P-Space, we introduce a matrix orthogonalization constraint to ensure the independence of global prototypes. Finally, we conduct extensive experiments in PTaRL coupled with state-of-the-art deep tabular ML models on various tabular benchmarks and the results have shown our consistent superiority.
Boosting Protein Language Models with Negative Sample Mining
May 28, 2024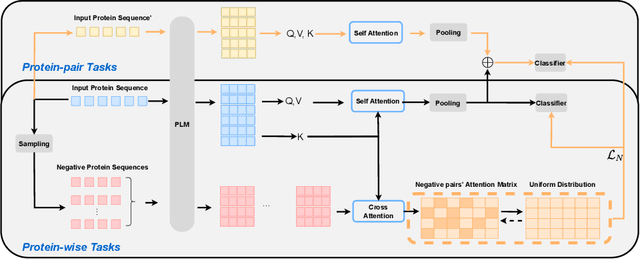
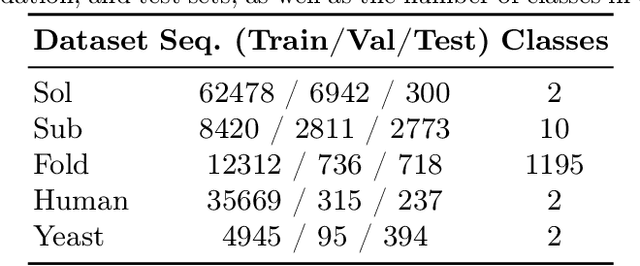
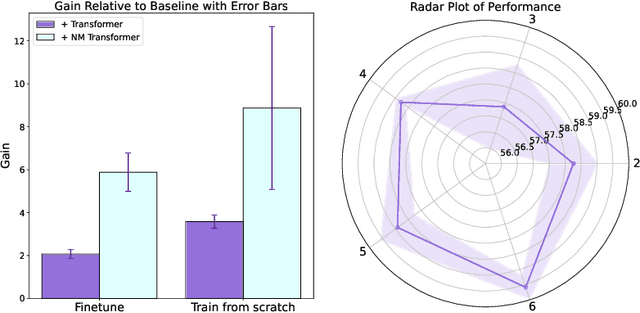
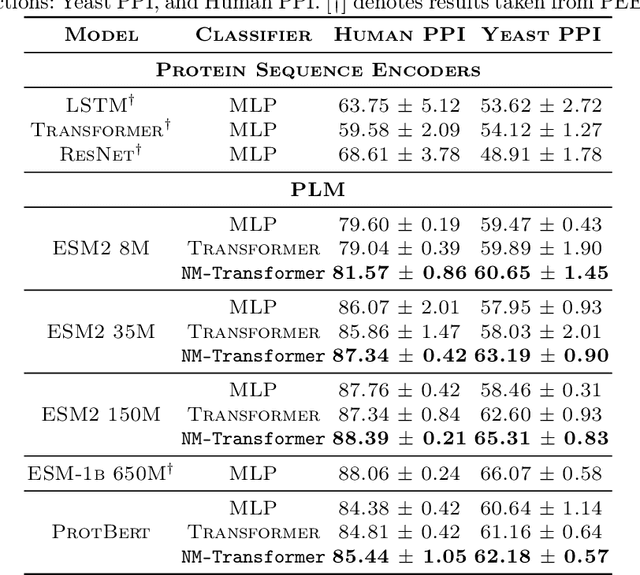
Abstract:We introduce a pioneering methodology for boosting large language models in the domain of protein representation learning. Our primary contribution lies in the refinement process for correlating the over-reliance on co-evolution knowledge, in a way that networks are trained to distill invaluable insights from negative samples, constituted by protein pairs sourced from disparate categories. By capitalizing on this novel approach, our technique steers the training of transformer-based models within the attention score space. This advanced strategy not only amplifies performance but also reflects the nuanced biological behaviors exhibited by proteins, offering aligned evidence with traditional biological mechanisms such as protein-protein interaction. We experimentally observed improved performance on various tasks over datasets, on top of several well-established large protein models. This innovative paradigm opens up promising horizons for further progress in the realms of protein research and computational biology.
Traffic Prediction with Transfer Learning: A Mutual Information-based Approach
Mar 13, 2023



Abstract:In modern traffic management, one of the most essential yet challenging tasks is accurately and timely predicting traffic. It has been well investigated and examined that deep learning-based Spatio-temporal models have an edge when exploiting Spatio-temporal relationships in traffic data. Typically, data-driven models require vast volumes of data, but gathering data in small cities can be difficult owing to constraints such as equipment deployment and maintenance costs. To resolve this problem, we propose TrafficTL, a cross-city traffic prediction approach that uses big data from other cities to aid data-scarce cities in traffic prediction. Utilizing a periodicity-based transfer paradigm, it identifies data similarity and reduces negative transfer caused by the disparity between two data distributions from distant cities. In addition, the suggested method employs graph reconstruction techniques to rectify defects in data from small data cities. TrafficTL is evaluated by comprehensive case studies on three real-world datasets and outperforms the state-of-the-art baseline by around 8 to 25 percent.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge