Yaoyao Xu
Enhancing Graph Self-Supervised Learning with Graph Interplay
Oct 08, 2024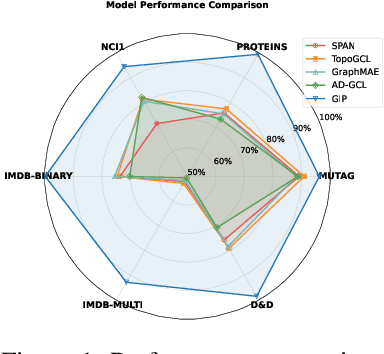
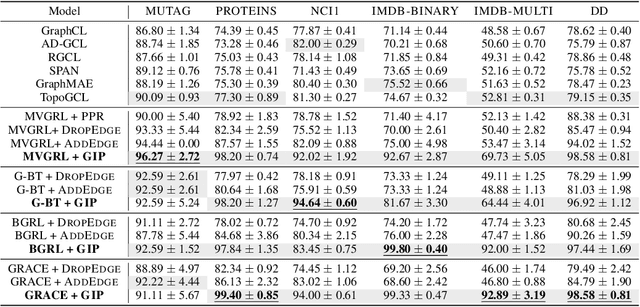
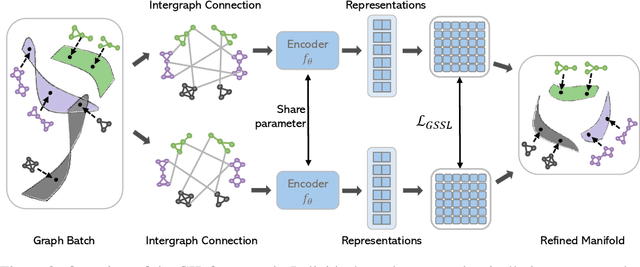
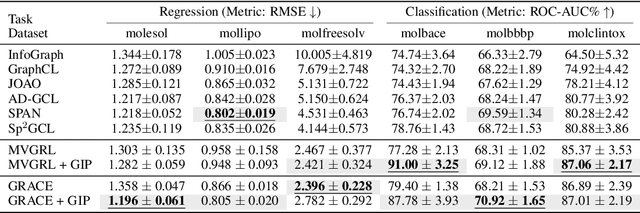
Abstract:Graph self-supervised learning (GSSL) has emerged as a compelling framework for extracting informative representations from graph-structured data without extensive reliance on labeled inputs. In this study, we introduce Graph Interplay (GIP), an innovative and versatile approach that significantly enhances the performance equipped with various existing GSSL methods. To this end, GIP advocates direct graph-level communications by introducing random inter-graph edges within standard batches. Against GIP's simplicity, we further theoretically show that \textsc{GIP} essentially performs a principled manifold separation via combining inter-graph message passing and GSSL, bringing about more structured embedding manifolds and thus benefits a series of downstream tasks. Our empirical study demonstrates that GIP surpasses the performance of prevailing GSSL methods across multiple benchmarks by significant margins, highlighting its potential as a breakthrough approach. Besides, GIP can be readily integrated into a series of GSSL methods and consistently offers additional performance gain. This advancement not only amplifies the capability of GSSL but also potentially sets the stage for a novel graph learning paradigm in a broader sense.
Do spectral cues matter in contrast-based graph self-supervised learning?
May 30, 2024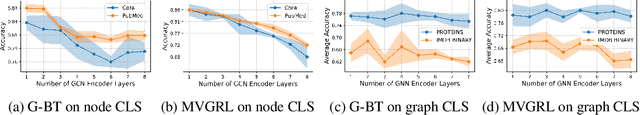
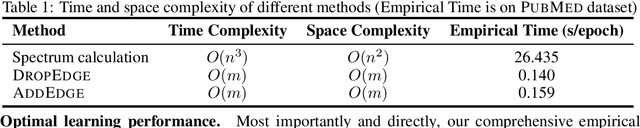
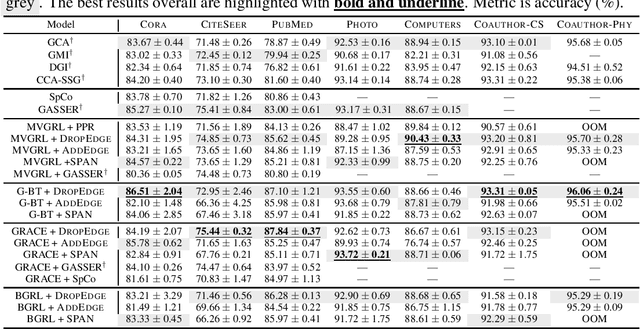
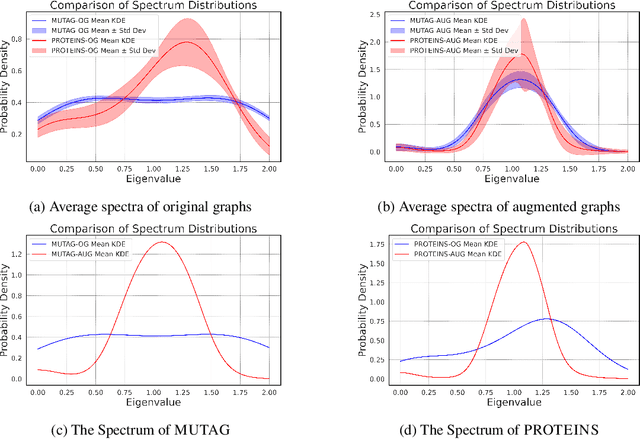
Abstract:The recent surge in contrast-based graph self-supervised learning has prominently featured an intensified exploration of spectral cues. However, an intriguing paradox emerges, as methods grounded in seemingly conflicting assumptions or heuristic approaches regarding the spectral domain demonstrate notable enhancements in learning performance. This paradox prompts a critical inquiry into the genuine contribution of spectral information to contrast-based graph self-supervised learning. This study undertakes an extensive investigation into this inquiry, conducting a thorough study of the relationship between spectral characteristics and the learning outcomes of contemporary methodologies. Based on this analysis, we claim that the effectiveness and significance of spectral information need to be questioned. Instead, we revisit simple edge perturbation: random edge dropping designed for node-level self-supervised learning and random edge adding intended for graph-level self-supervised learning. Compelling evidence is presented that these simple yet effective strategies consistently yield superior performance while demanding significantly fewer computational resources compared to all prior spectral augmentation methods. The proposed insights represent a significant leap forward in the field, potentially reshaping the understanding and implementation of graph self-supervised learning.
Boosting Protein Language Models with Negative Sample Mining
May 28, 2024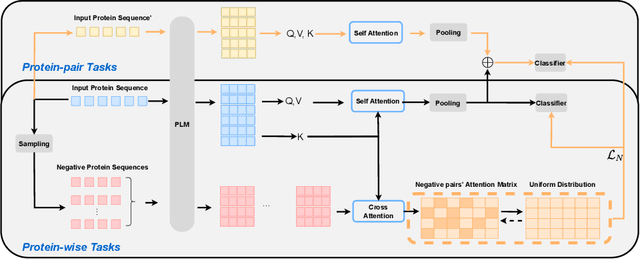
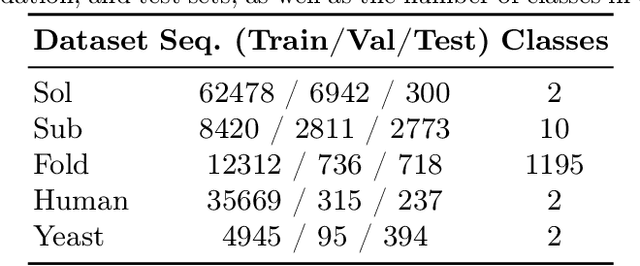
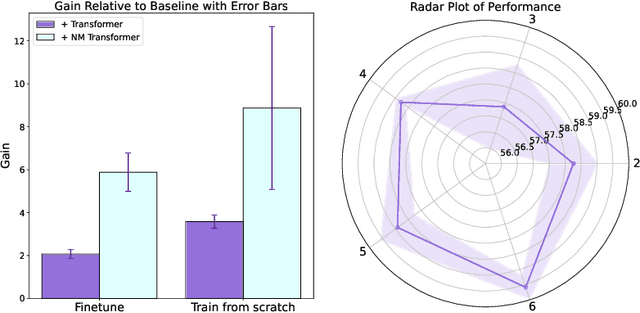
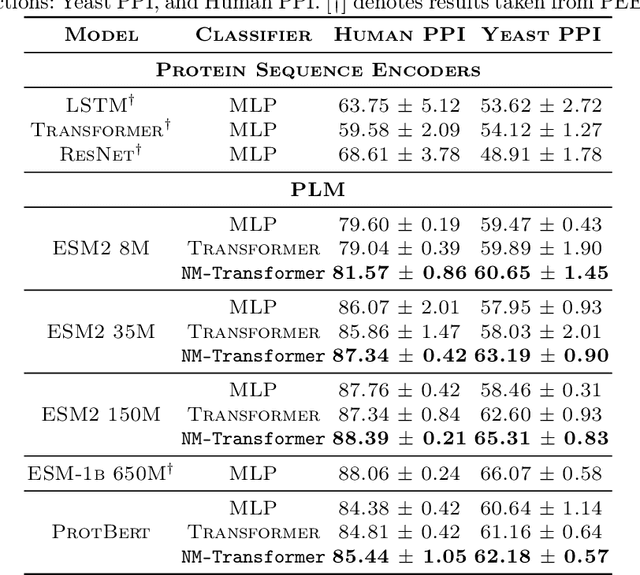
Abstract:We introduce a pioneering methodology for boosting large language models in the domain of protein representation learning. Our primary contribution lies in the refinement process for correlating the over-reliance on co-evolution knowledge, in a way that networks are trained to distill invaluable insights from negative samples, constituted by protein pairs sourced from disparate categories. By capitalizing on this novel approach, our technique steers the training of transformer-based models within the attention score space. This advanced strategy not only amplifies performance but also reflects the nuanced biological behaviors exhibited by proteins, offering aligned evidence with traditional biological mechanisms such as protein-protein interaction. We experimentally observed improved performance on various tasks over datasets, on top of several well-established large protein models. This innovative paradigm opens up promising horizons for further progress in the realms of protein research and computational biology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge