Boosting Protein Language Models with Negative Sample Mining
Paper and Code
May 28, 2024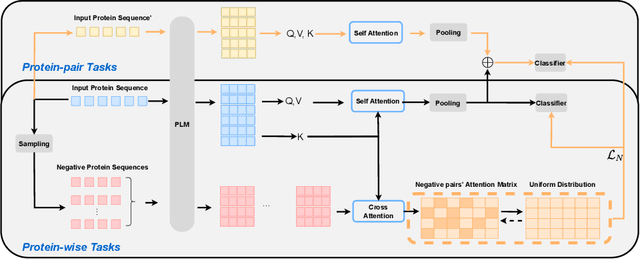
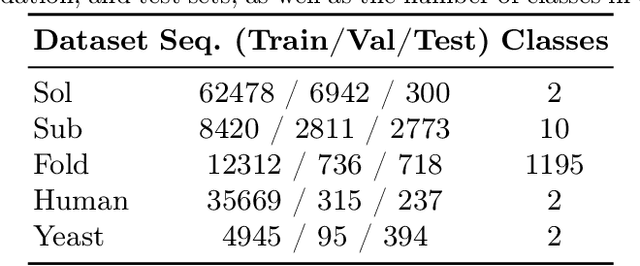
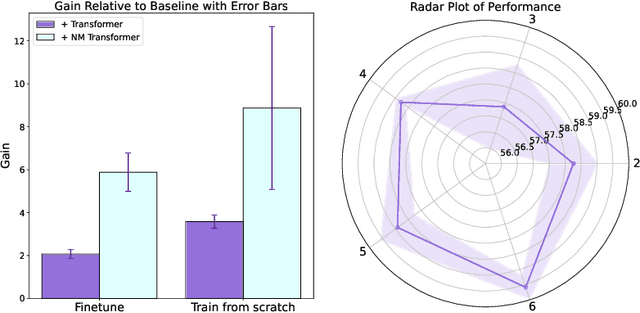
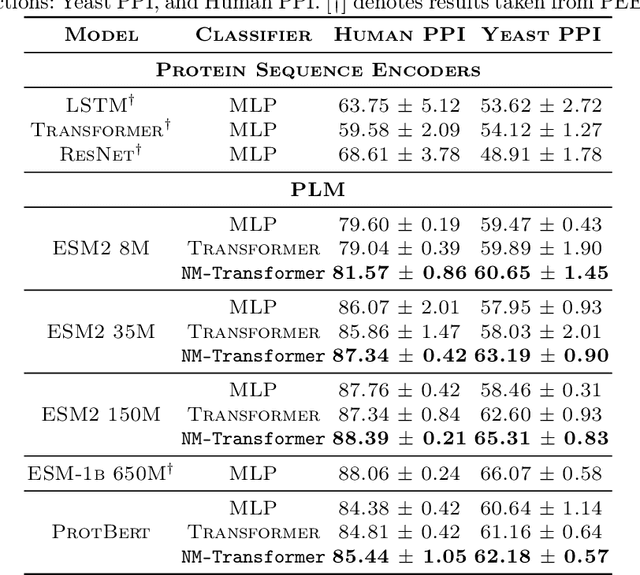
We introduce a pioneering methodology for boosting large language models in the domain of protein representation learning. Our primary contribution lies in the refinement process for correlating the over-reliance on co-evolution knowledge, in a way that networks are trained to distill invaluable insights from negative samples, constituted by protein pairs sourced from disparate categories. By capitalizing on this novel approach, our technique steers the training of transformer-based models within the attention score space. This advanced strategy not only amplifies performance but also reflects the nuanced biological behaviors exhibited by proteins, offering aligned evidence with traditional biological mechanisms such as protein-protein interaction. We experimentally observed improved performance on various tasks over datasets, on top of several well-established large protein models. This innovative paradigm opens up promising horizons for further progress in the realms of protein research and computational biology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge