Timothy Atkinson
Overconfident Oracles: Limitations of In Silico Sequence Design Benchmarking
Feb 24, 2025Abstract:Machine learning methods can automate the in silico design of biological sequences, aiming to reduce costs and accelerate medical research. Given the limited access to wet labs, in silico design methods commonly use an oracle model to evaluate de novo generated sequences. However, the use of different oracle models across methods makes it challenging to compare them reliably, motivating the question: are in silico sequence design benchmarks reliable? In this work, we examine 12 sequence design methods that utilise ML oracles common in the literature and find that there are significant challenges with their cross-consistency and reproducibility. Indeed, oracles differing by architecture, or even just training seed, are shown to yield conflicting relative performance with our analysis suggesting poor out-of-distribution generalisation as a key issue. To address these challenges, we propose supplementing the evaluation with a suite of biophysical measures to assess the viability of generated sequences and limit out-of-distribution sequences the oracle is required to score, thereby improving the robustness of the design procedure. Our work aims to highlight potential pitfalls in the current evaluation process and contribute to the development of robust benchmarks, ultimately driving the improvement of in silico design methods.
Learning the Language of Protein Structure
May 24, 2024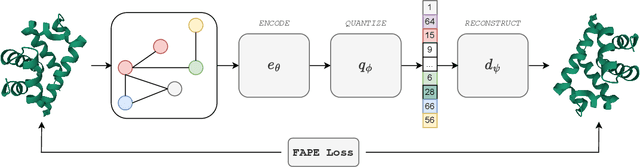
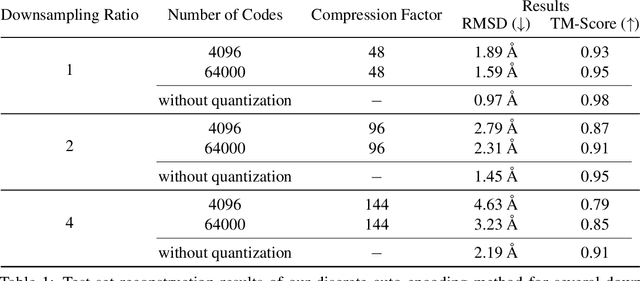
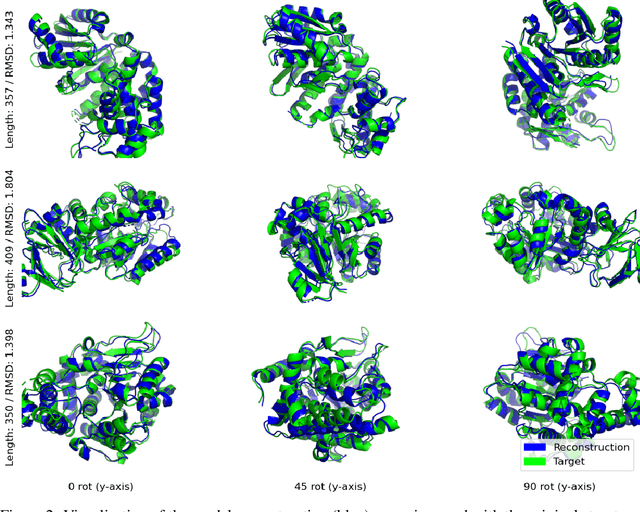
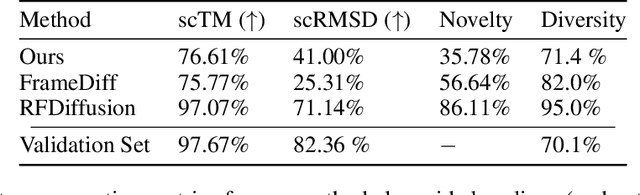
Abstract:Representation learning and \emph{de novo} generation of proteins are pivotal computational biology tasks. Whilst natural language processing (NLP) techniques have proven highly effective for protein sequence modelling, structure modelling presents a complex challenge, primarily due to its continuous and three-dimensional nature. Motivated by this discrepancy, we introduce an approach using a vector-quantized autoencoder that effectively tokenizes protein structures into discrete representations. This method transforms the continuous, complex space of protein structures into a manageable, discrete format with a codebook ranging from 4096 to 64000 tokens, achieving high-fidelity reconstructions with backbone root mean square deviations (RMSD) of approximately 1-5 \AA. To demonstrate the efficacy of our learned representations, we show that a simple GPT model trained on our codebooks can generate novel, diverse, and designable protein structures. Our approach not only provides representations of protein structure, but also mitigates the challenges of disparate modal representations and sets a foundation for seamless, multi-modal integration, enhancing the capabilities of computational methods in protein design.
Bayesian Flow Networks
Aug 14, 2023



Abstract:This paper introduces Bayesian Flow Networks (BFNs), a new class of generative model in which the parameters of a set of independent distributions are modified with Bayesian inference in the light of noisy data samples, then passed as input to a neural network that outputs a second, interdependent distribution. Starting from a simple prior and iteratively updating the two distributions yields a generative procedure similar to the reverse process of diffusion models; however it is conceptually simpler in that no forward process is required. Discrete and continuous-time loss functions are derived for continuous, discretised and discrete data, along with sample generation procedures. Notably, the network inputs for discrete data lie on the probability simplex, and are therefore natively differentiable, paving the way for gradient-based sample guidance and few-step generation in discrete domains such as language modelling. The loss function directly optimises data compression and places no restrictions on the network architecture. In our experiments BFNs achieve competitive log-likelihoods for image modelling on dynamically binarized MNIST and CIFAR-10, and outperform all known discrete diffusion models on the text8 character-level language modelling task.
Tiny Classifier Circuits: Evolving Accelerators for Tabular Data
Feb 28, 2023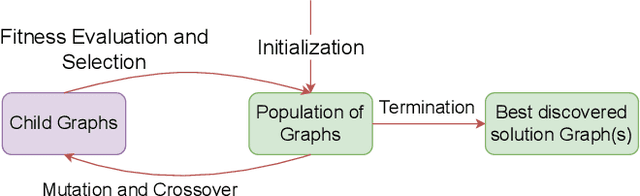
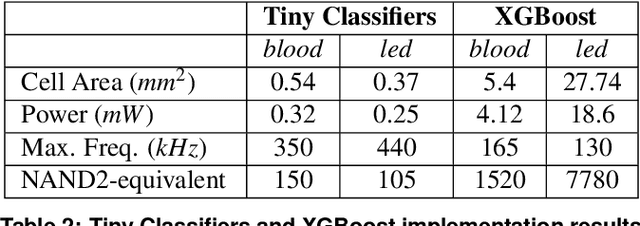
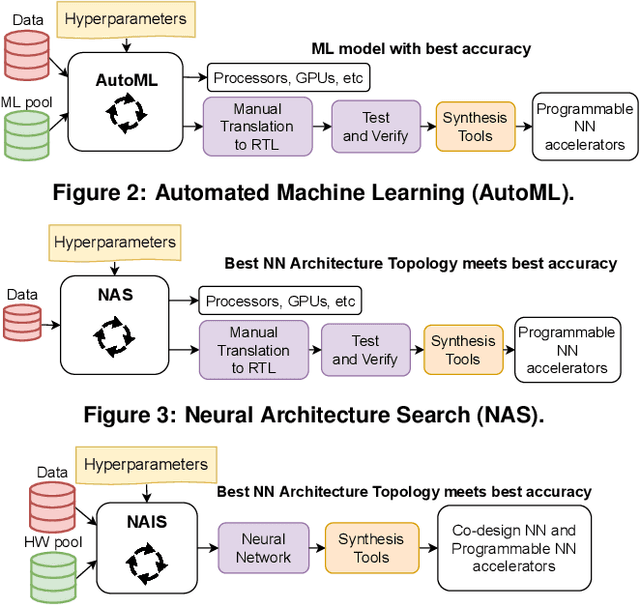
Abstract:A typical machine learning (ML) development cycle for edge computing is to maximise the performance during model training and then minimise the memory/area footprint of the trained model for deployment on edge devices targeting CPUs, GPUs, microcontrollers, or custom hardware accelerators. This paper proposes a methodology for automatically generating predictor circuits for classification of tabular data with comparable prediction performance to conventional ML techniques while using substantially fewer hardware resources and power. The proposed methodology uses an evolutionary algorithm to search over the space of logic gates and automatically generates a classifier circuit with maximised training prediction accuracy. Classifier circuits are so tiny (i.e., consisting of no more than 300 logic gates) that they are called "Tiny Classifier" circuits, and can efficiently be implemented in ASIC or on an FPGA. We empirically evaluate the automatic Tiny Classifier circuit generation methodology or "Auto Tiny Classifiers" on a wide range of tabular datasets, and compare it against conventional ML techniques such as Amazon's AutoGluon, Google's TabNet and a neural search over Multi-Layer Perceptrons. Despite Tiny Classifiers being constrained to a few hundred logic gates, we observe no statistically significant difference in prediction performance in comparison to the best-performing ML baseline. When synthesised as a Silicon chip, Tiny Classifiers use 8-56x less area and 4-22x less power. When implemented as an ultra-low cost chip on a flexible substrate (i.e., FlexIC), they occupy 10-75x less area and consume 13-75x less power compared to the most hardware-efficient ML baseline. On an FPGA, Tiny Classifiers consume 3-11x fewer resources.
EvoTorch: Scalable Evolutionary Computation in Python
Feb 27, 2023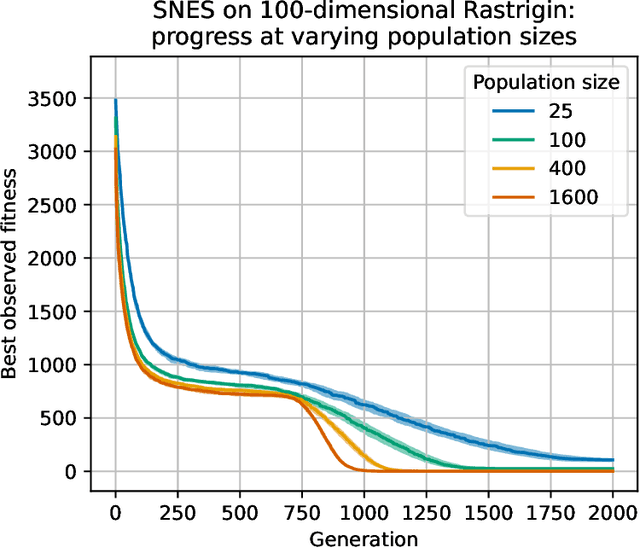
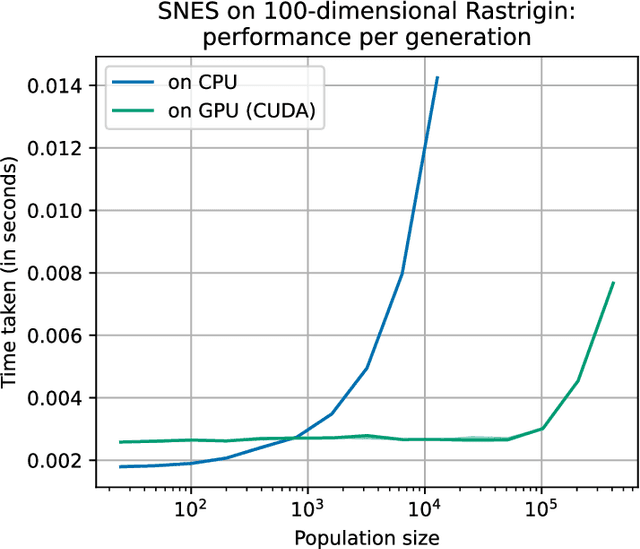
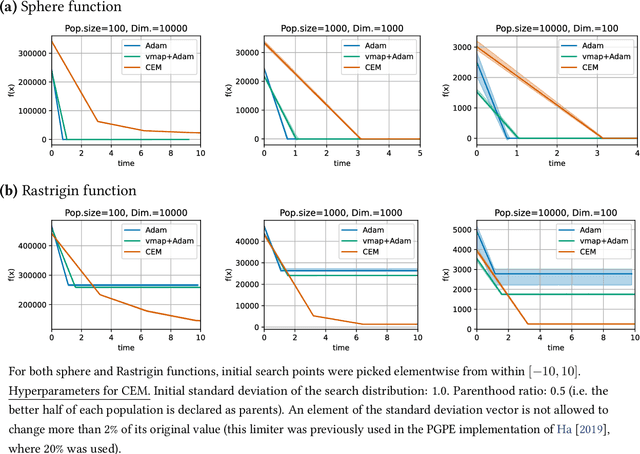
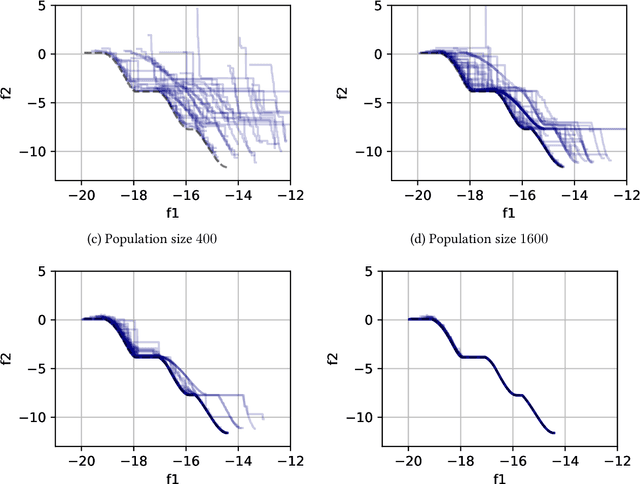
Abstract:Evolutionary computation is an important component within various fields such as artificial intelligence research, reinforcement learning, robotics, industrial automation and/or optimization, engineering design, etc. Considering the increasing computational demands and the dimensionalities of modern optimization problems, the requirement for scalable, re-usable, and practical evolutionary algorithm implementations has been growing. To address this requirement, we present EvoTorch: an evolutionary computation library designed to work with high-dimensional optimization problems, with GPU support and with high parallelization capabilities. EvoTorch is based on and seamlessly works with the PyTorch library, and therefore, allows the users to define their optimization problems using a well-known API.
Automatic design of novel potential 3CL$^{\text{pro}}$ and PL$^{\text{pro}}$ inhibitors
Jan 29, 2021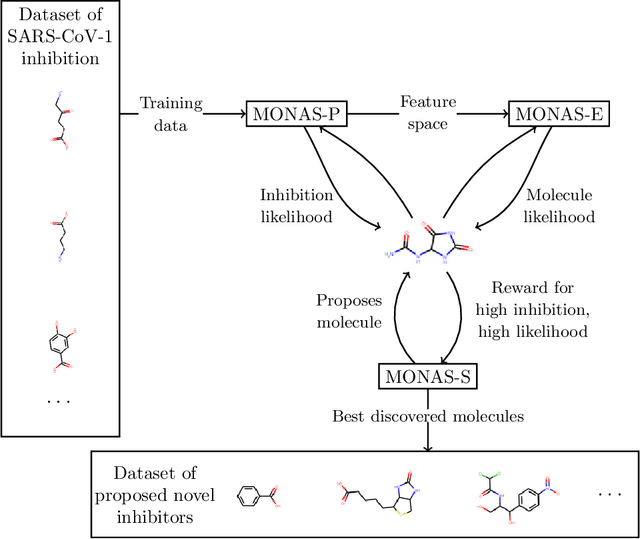
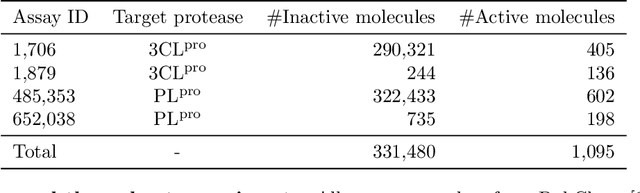
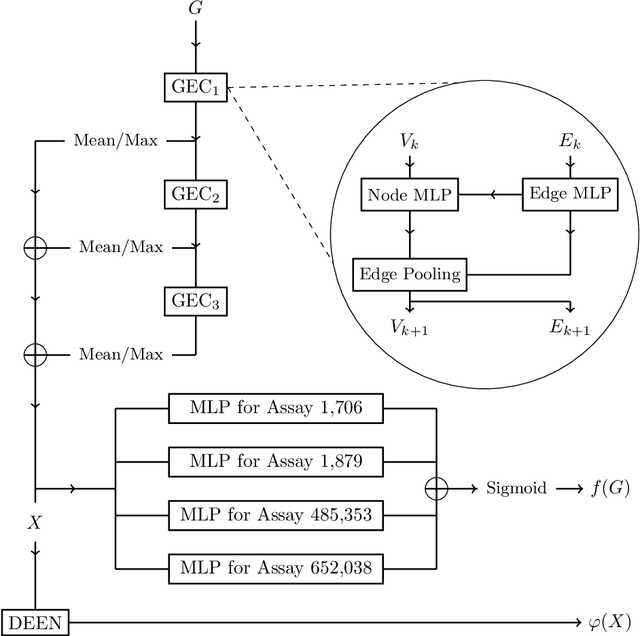
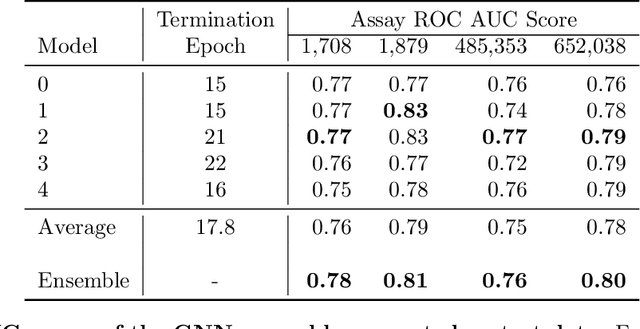
Abstract:With the goal of designing novel inhibitors for SARS-CoV-1 and SARS-CoV-2, we propose the general molecule optimization framework, Molecular Neural Assay Search (MONAS), consisting of three components: a property predictor which identifies molecules with specific desirable properties, an energy model which approximates the statistical similarity of a given molecule to known training molecules, and a molecule search method. In this work, these components are instantiated with graph neural networks (GNNs), Deep Energy Estimator Networks (DEEN) and Monte Carlo tree search (MCTS), respectively. This implementation is used to identify 120K molecules (out of 40-million explored) which the GNN determined to be likely SARS-CoV-1 inhibitors, and, at the same time, are statistically close to the dataset used to train the GNN.
Semantic Neutral Drift
Oct 24, 2018



Abstract:We introduce the concept of Semantic Neutral Drift (SND) for evolutionary algorithms, where we exploit equivalence laws to design semantics preserving mutations guaranteed to preserve individuals' fitness scores. A number of digital circuit benchmark problems have been implemented with rule-based graph programs and empirically evaluated, demonstrating quantitative improvements in evolutionary performance. Analysis reveals that the benefits of the designed SND reside in more complex processes than simple growth of individuals, and that there are circumstances where it is beneficial to choose otherwise detrimental parameters for an evolutionary algorithm if that facilitates the inclusion of SND.
The Text-Based Adventure AI Competition
Oct 19, 2018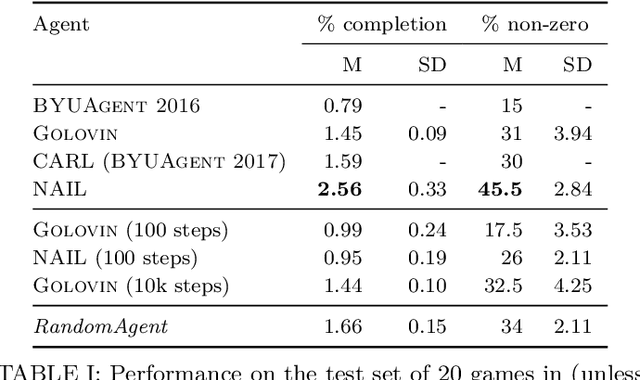
Abstract:In 2016, 2017, and 2018 at the IEEE Conference on Computational Intelligence in Games, the authors of this paper ran a competition for agents that can play classic text-based adventure games. This competition fills a gap in existing game AI competitions that have typically focussed on traditional card/board games or modern video games with graphical interfaces. By providing a platform for evaluating agents in text-based adventures, the competition provides a novel benchmark for game AI with unique challenges for natural language understanding and generation. This paper summarises the three competitions ran in 2016, 2017, and 2018 (including details of open source implementations of both the competition framework and our competitors) and presents the results of an improved evaluation of these competitors across 20 games.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge