Théo Estienne
Université Paris-Saclay, CentraleSupélec, Mathématiques et Informatique pour la Complexité et les Systèmes, Inria Saclay, Université Paris-Saclay, Institut Gustave Roussy, Inserm, Radiothérapie Moléculaire et Innovation Thérapeutique
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021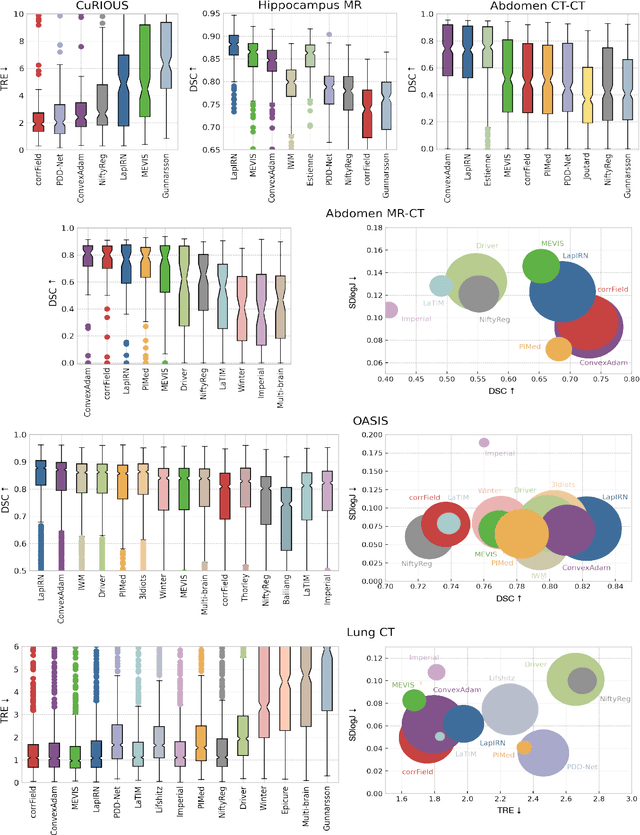
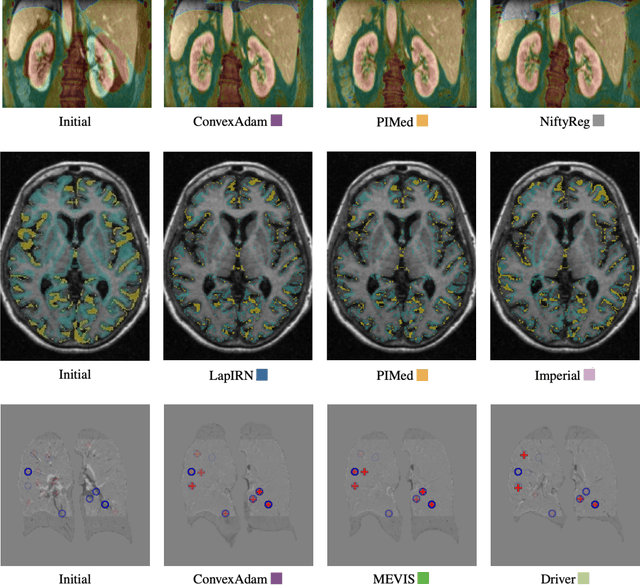
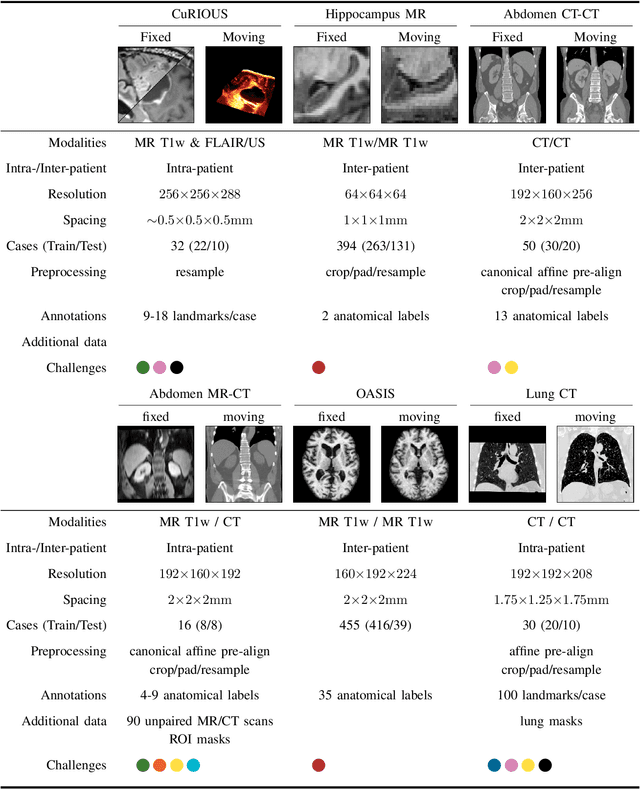
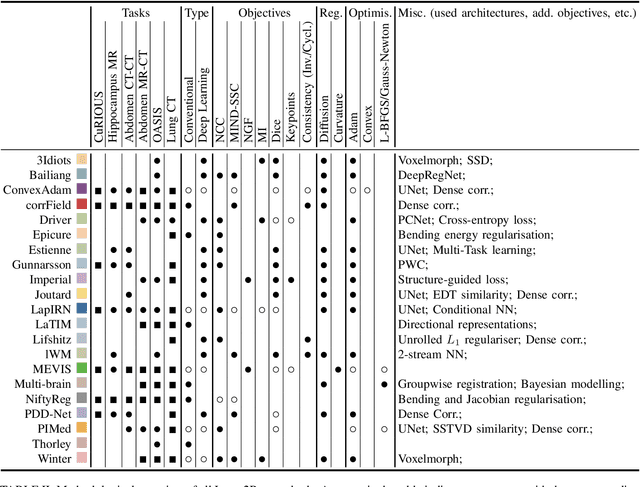
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
MICS : Multi-steps, Inverse Consistency and Symmetric deep learning registration network
Nov 23, 2021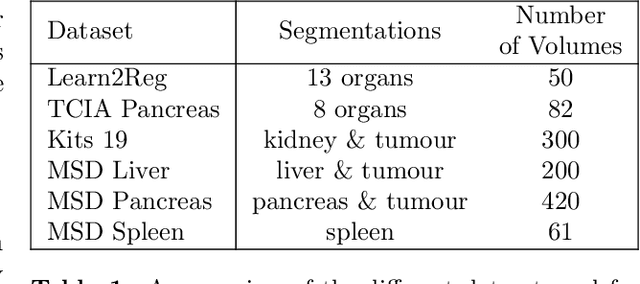
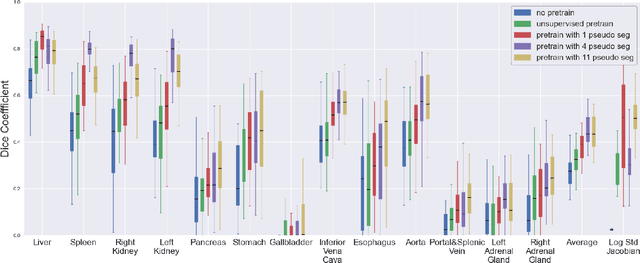
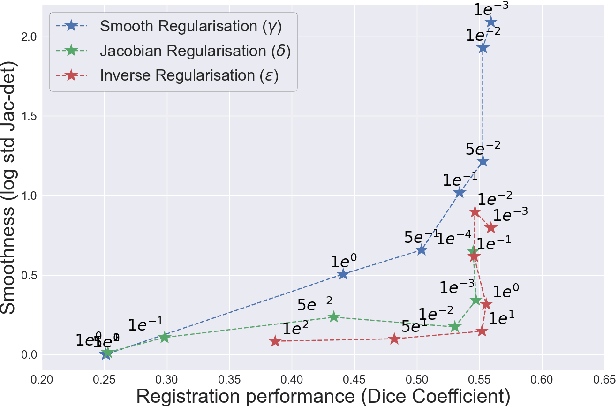

Abstract:Deformable registration consists of finding the best dense correspondence between two different images. Many algorithms have been published, but the clinical application was made difficult by the high calculation time needed to solve the optimisation problem. Deep learning overtook this limitation by taking advantage of GPU calculation and the learning process. However, many deep learning methods do not take into account desirable properties respected by classical algorithms. In this paper, we present MICS, a novel deep learning algorithm for medical imaging registration. As registration is an ill-posed problem, we focused our algorithm on the respect of different properties: inverse consistency, symmetry and orientation conservation. We also combined our algorithm with a multi-step strategy to refine and improve the deformation grid. While many approaches applied registration to brain MRI, we explored a more challenging body localisation: abdominal CT. Finally, we evaluated our method on a dataset used during the Learn2Reg challenge, allowing a fair comparison with published methods.
Exploring Deep Registration Latent Spaces
Jul 23, 2021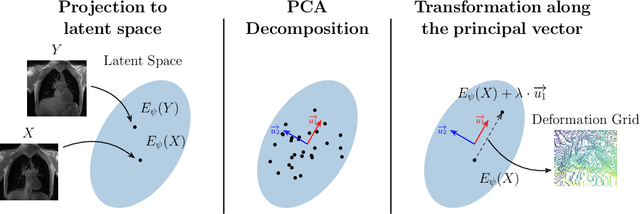
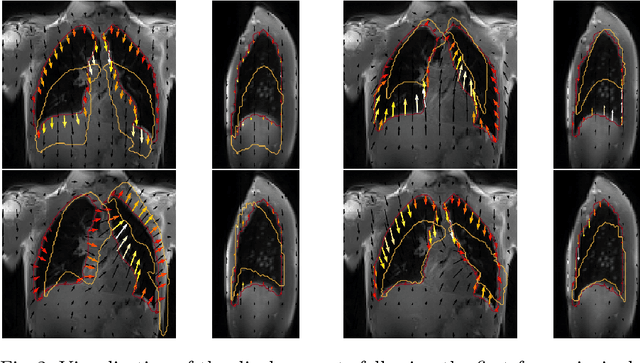
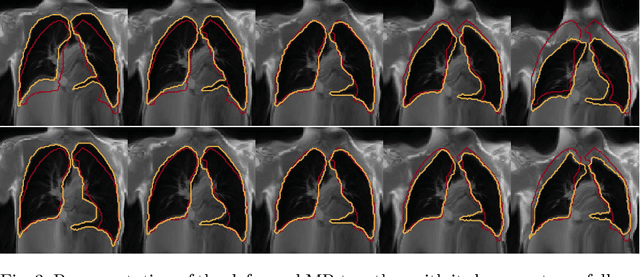
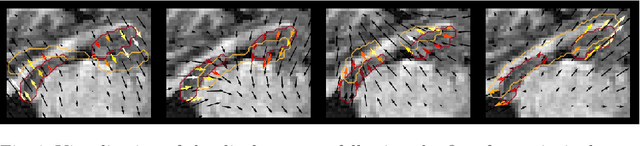
Abstract:Explainability of deep neural networks is one of the most challenging and interesting problems in the field. In this study, we investigate the topic focusing on the interpretability of deep learning-based registration methods. In particular, with the appropriate model architecture and using a simple linear projection, we decompose the encoding space, generating a new basis, and we empirically show that this basis captures various decomposed anatomically aware geometrical transformations. We perform experiments using two different datasets focusing on lungs and hippocampus MRI. We show that such an approach can decompose the highly convoluted latent spaces of registration pipelines in an orthogonal space with several interesting properties. We hope that this work could shed some light on a better understanding of deep learning-based registration methods.
Weakly supervised pan-cancer segmentation tool
May 10, 2021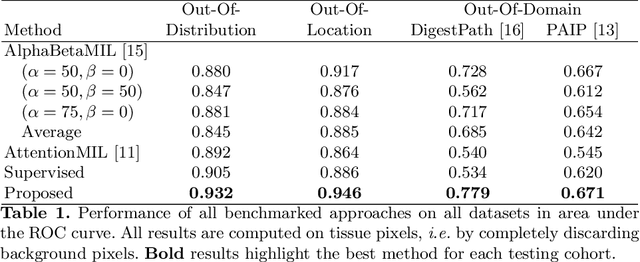
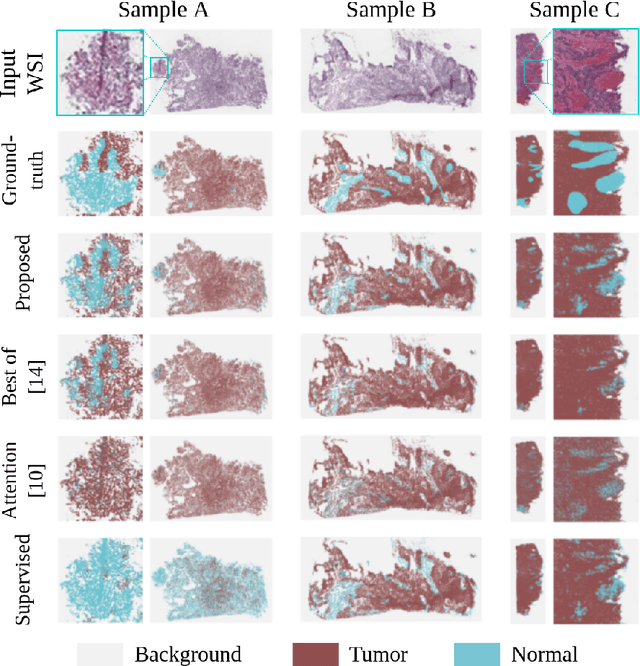
Abstract:The vast majority of semantic segmentation approaches rely on pixel-level annotations that are tedious and time consuming to obtain and suffer from significant inter and intra-expert variability. To address these issues, recent approaches have leveraged categorical annotations at the slide-level, that in general suffer from robustness and generalization. In this paper, we propose a novel weakly supervised multi-instance learning approach that deciphers quantitative slide-level annotations which are fast to obtain and regularly present in clinical routine. The extreme potentials of the proposed approach are demonstrated for tumor segmentation of solid cancer subtypes. The proposed approach achieves superior performance in out-of-distribution, out-of-location, and out-of-domain testing sets.
Cancer Gene Profiling through Unsupervised Discovery
Feb 11, 2021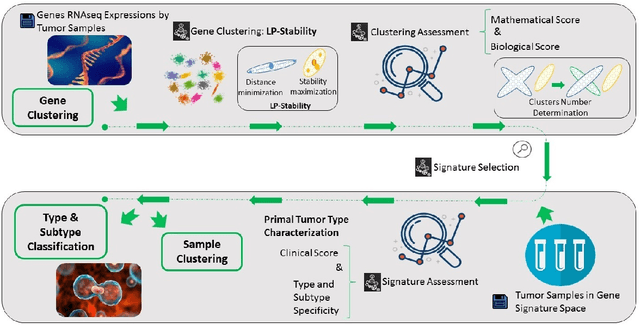
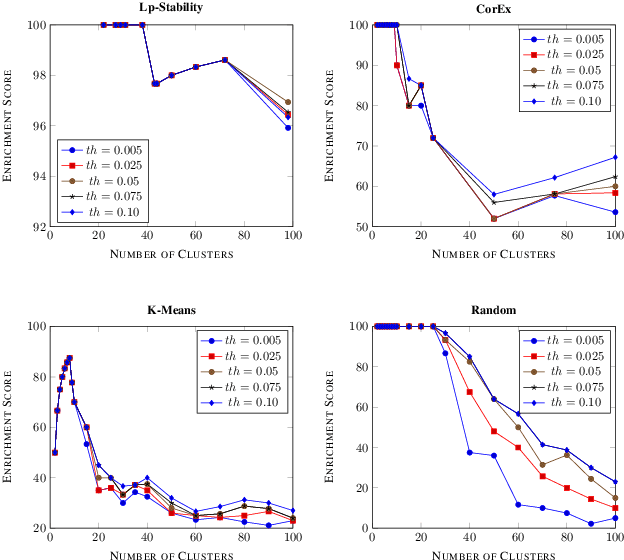
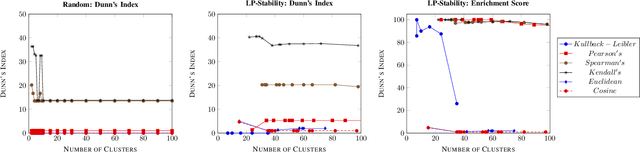
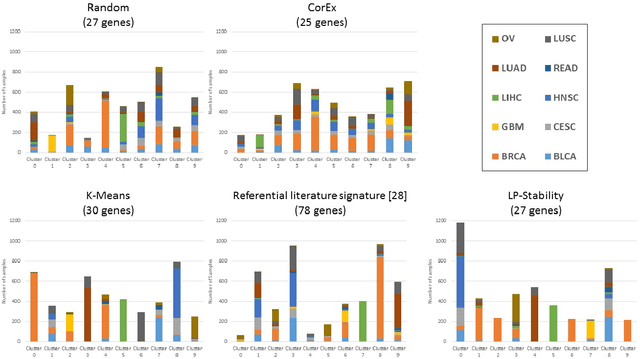
Abstract:Precision medicine is a paradigm shift in healthcare relying heavily on genomics data. However, the complexity of biological interactions, the large number of genes as well as the lack of comparisons on the analysis of data, remain a tremendous bottleneck regarding clinical adoption. In this paper, we introduce a novel, automatic and unsupervised framework to discover low-dimensional gene biomarkers. Our method is based on the LP-Stability algorithm, a high dimensional center-based unsupervised clustering algorithm, that offers modularity as concerns metric functions and scalability, while being able to automatically determine the best number of clusters. Our evaluation includes both mathematical and biological criteria. The recovered signature is applied to a variety of biological tasks, including screening of biological pathways and functions, and characterization relevance on tumor types and subtypes. Quantitative comparisons among different distance metrics, commonly used clustering methods and a referential gene signature used in the literature, confirm state of the art performance of our approach. In particular, our signature, that is based on 27 genes, reports at least $30$ times better mathematical significance (average Dunn's Index) and 25% better biological significance (average Enrichment in Protein-Protein Interaction) than those produced by other referential clustering methods. Finally, our signature reports promising results on distinguishing immune inflammatory and immune desert tumors, while reporting a high balanced accuracy of 92% on tumor types classification and averaged balanced accuracy of 68% on tumor subtypes classification, which represents, respectively 7% and 9% higher performance compared to the referential signature.
Deep learning based registration using spatial gradients and noisy segmentation labels
Oct 21, 2020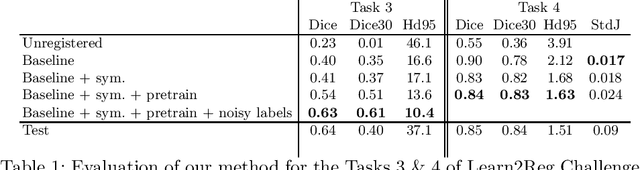
Abstract:Image registration is one of the most challenging problems in medical image analysis. In the recent years, deep learning based approaches became quite popular, providing fast and performing registration strategies. In this short paper, we summarise our work presented on Learn2Reg challenge 2020. The main contributions of our work rely on (i) a symmetric formulation, predicting the transformations from source to target and from target to source simultaneously, enforcing the trained representations to be similar and (ii) integration of variety of publicly available datasets used both for pretraining and for augmenting segmentation labels. Our method reports a mean dice of $0.64$ for task 3 and $0.85$ for task 4 on the test sets, taking third place on the challenge. Our code and models are publicly available at https://github.com/TheoEst/abdominal_registration and \https://github.com/TheoEst/hippocampus_registration.
Weakly supervised multiple instance learning histopathological tumor segmentation
Apr 21, 2020
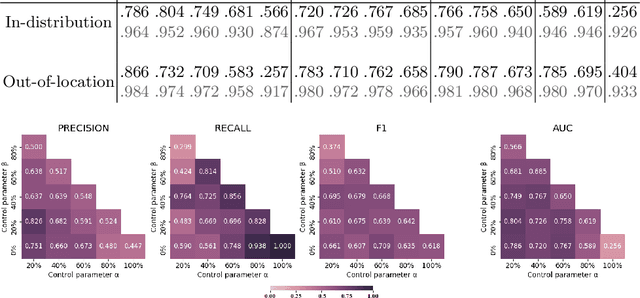
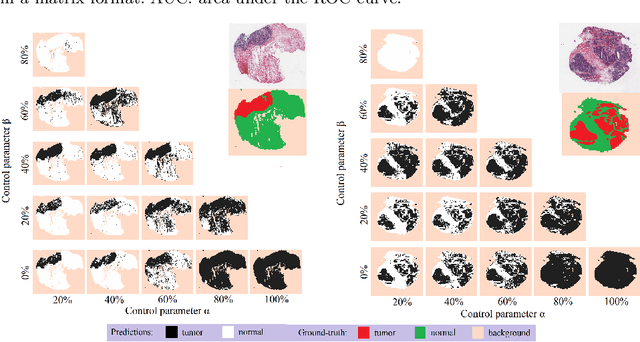
Abstract:Histopathological image segmentation is a challenging and important topic in medical imaging with tremendous potential impact in clinical practice. State of the art methods relying on hand-crafted annotations that reduce the scope of the solutions since digital histology suffers from standardization and samples differ significantly between cancer phenotypes. To this end, in this paper, we propose a weakly supervised framework relying on weak standard clinical practice annotations, available in most medical centers. In particular, we exploit a multiple instance learning scheme providing a label for each instance, establishing a detailed segmentation of whole slide images. The potential of the framework is assessed with multi-centric data experiments using The Cancer Genome Atlas repository and the publicly available PatchCamelyon dataset. Promising results when compared with experts' annotations demonstrate the potentials of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge