Charlotte Robert
Université Paris-Saclay, Institut Gustave Roussy, Inserm, Radiothérapie Moléculaire et Innovation Thérapeutique
GuidedRec: Guiding Ill-Posed Unsupervised Volumetric Recovery
May 20, 2024



Abstract:We introduce a novel unsupervised approach to reconstructing a 3D volume from only two planar projections that exploits a previous\-ly-captured 3D volume of the patient. Such volume is readily available in many important medical procedures and previous methods already used such a volume. Earlier methods that work by deforming this volume to match the projections typically fail when the number of projections is very low as the alignment becomes underconstrained. We show how to use a generative model of the volume structures to constrain the deformation and obtain a correct estimate. Moreover, our method is not bounded to a specific sensor calibration and can be applied to new calibrations without retraining. We evaluate our approach on a challenging dataset and show it outperforms state-of-the-art methods. As a result, our method could be used in treatment scenarios such as surgery and radiotherapy while drastically reducing patient radiation exposure.
Cancer Gene Profiling through Unsupervised Discovery
Feb 11, 2021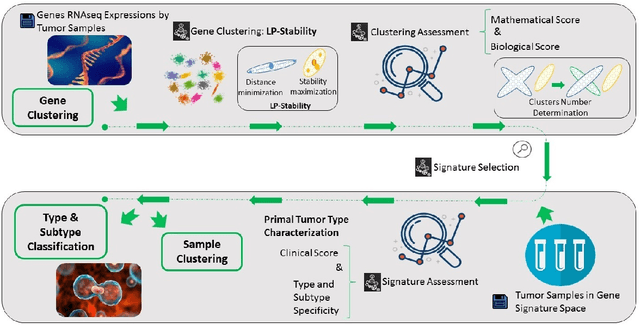
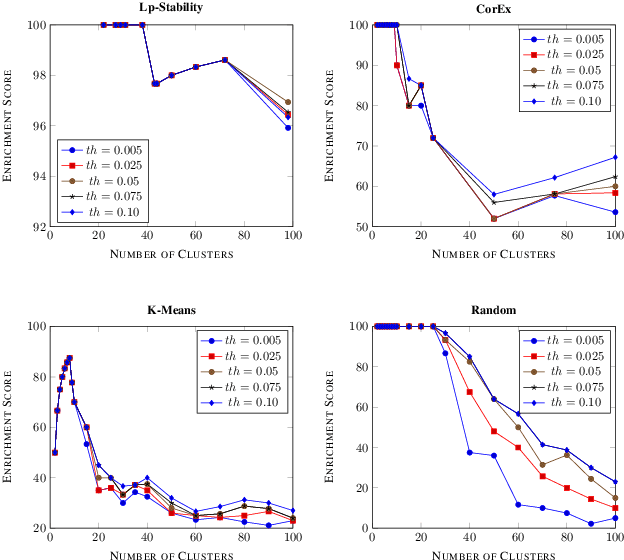
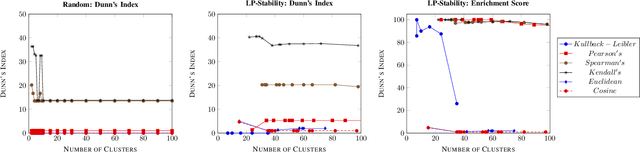
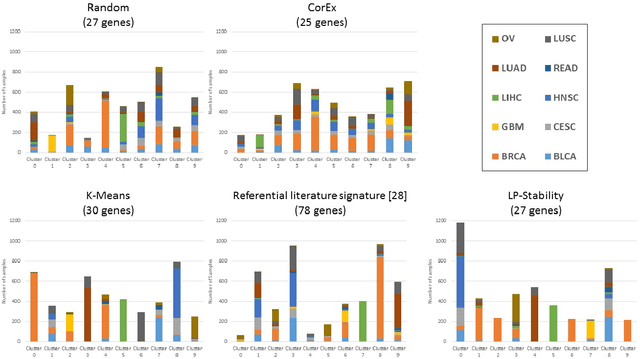
Abstract:Precision medicine is a paradigm shift in healthcare relying heavily on genomics data. However, the complexity of biological interactions, the large number of genes as well as the lack of comparisons on the analysis of data, remain a tremendous bottleneck regarding clinical adoption. In this paper, we introduce a novel, automatic and unsupervised framework to discover low-dimensional gene biomarkers. Our method is based on the LP-Stability algorithm, a high dimensional center-based unsupervised clustering algorithm, that offers modularity as concerns metric functions and scalability, while being able to automatically determine the best number of clusters. Our evaluation includes both mathematical and biological criteria. The recovered signature is applied to a variety of biological tasks, including screening of biological pathways and functions, and characterization relevance on tumor types and subtypes. Quantitative comparisons among different distance metrics, commonly used clustering methods and a referential gene signature used in the literature, confirm state of the art performance of our approach. In particular, our signature, that is based on 27 genes, reports at least $30$ times better mathematical significance (average Dunn's Index) and 25% better biological significance (average Enrichment in Protein-Protein Interaction) than those produced by other referential clustering methods. Finally, our signature reports promising results on distinguishing immune inflammatory and immune desert tumors, while reporting a high balanced accuracy of 92% on tumor types classification and averaged balanced accuracy of 68% on tumor subtypes classification, which represents, respectively 7% and 9% higher performance compared to the referential signature.
Top 10 BraTS 2020 challenge solution: Brain tumor segmentation with self-ensembled, deeply-supervised 3D-Unet like neural networks
Oct 30, 2020

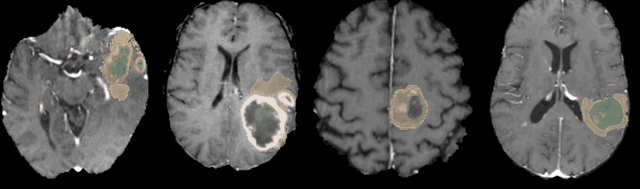

Abstract:Brain tumor segmentation is a critical task for patient's disease management. To this end, we trained multiple U-net like neural networks, mainly with deep supervision and stochastic weight averaging, on the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2020 training dataset, in a cross-validated fashion. Final brain tumor segmentations were produced by first averaging independently two sets of models, and then custom merging the labelmaps to account for individual performance of each set. Our performance on the online validation dataset with test time augmentation were as follows: Dice of 0.81, 0.91 and 0.85; Hausdorff (95%) of 20.6, 4,3, 5.7 mm for the enhancing tumor, whole tumor and tumor core, respectively. Similarly, our ensemble achieved a Dice of 0.79, 0.89 and 0.84, as well as Hausdorff (95%) of 20.4, 6.7 and 19.5mm on the final test dataset. More complicated training schemes and neural network architectures were investigated, without significant performance gain, at the cost of greatly increased training time. While relatively straightforward, our approach yielded good and balanced performance for each tumor subregions. Our solution is open sourced at https://github.com/lescientifik/xxxxx.
Deep learning based registration using spatial gradients and noisy segmentation labels
Oct 21, 2020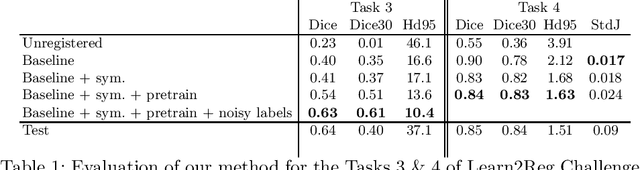
Abstract:Image registration is one of the most challenging problems in medical image analysis. In the recent years, deep learning based approaches became quite popular, providing fast and performing registration strategies. In this short paper, we summarise our work presented on Learn2Reg challenge 2020. The main contributions of our work rely on (i) a symmetric formulation, predicting the transformations from source to target and from target to source simultaneously, enforcing the trained representations to be similar and (ii) integration of variety of publicly available datasets used both for pretraining and for augmenting segmentation labels. Our method reports a mean dice of $0.64$ for task 3 and $0.85$ for task 4 on the test sets, taking third place on the challenge. Our code and models are publicly available at https://github.com/TheoEst/abdominal_registration and \https://github.com/TheoEst/hippocampus_registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge