Theophraste Henry
MICS : Multi-steps, Inverse Consistency and Symmetric deep learning registration network
Nov 23, 2021

Abstract:Deformable registration consists of finding the best dense correspondence between two different images. Many algorithms have been published, but the clinical application was made difficult by the high calculation time needed to solve the optimisation problem. Deep learning overtook this limitation by taking advantage of GPU calculation and the learning process. However, many deep learning methods do not take into account desirable properties respected by classical algorithms. In this paper, we present MICS, a novel deep learning algorithm for medical imaging registration. As registration is an ill-posed problem, we focused our algorithm on the respect of different properties: inverse consistency, symmetry and orientation conservation. We also combined our algorithm with a multi-step strategy to refine and improve the deformation grid. While many approaches applied registration to brain MRI, we explored a more challenging body localisation: abdominal CT. Finally, we evaluated our method on a dataset used during the Learn2Reg challenge, allowing a fair comparison with published methods.
Top 10 BraTS 2020 challenge solution: Brain tumor segmentation with self-ensembled, deeply-supervised 3D-Unet like neural networks
Oct 30, 2020

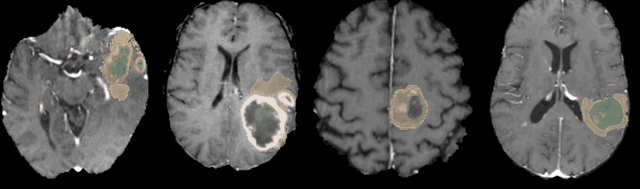

Abstract:Brain tumor segmentation is a critical task for patient's disease management. To this end, we trained multiple U-net like neural networks, mainly with deep supervision and stochastic weight averaging, on the Multimodal Brain Tumor Segmentation Challenge (BraTS) 2020 training dataset, in a cross-validated fashion. Final brain tumor segmentations were produced by first averaging independently two sets of models, and then custom merging the labelmaps to account for individual performance of each set. Our performance on the online validation dataset with test time augmentation were as follows: Dice of 0.81, 0.91 and 0.85; Hausdorff (95%) of 20.6, 4,3, 5.7 mm for the enhancing tumor, whole tumor and tumor core, respectively. Similarly, our ensemble achieved a Dice of 0.79, 0.89 and 0.84, as well as Hausdorff (95%) of 20.4, 6.7 and 19.5mm on the final test dataset. More complicated training schemes and neural network architectures were investigated, without significant performance gain, at the cost of greatly increased training time. While relatively straightforward, our approach yielded good and balanced performance for each tumor subregions. Our solution is open sourced at https://github.com/lescientifik/xxxxx.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge