Théophraste Henry
Université Paris-Saclay, Institut Gustave Roussy, Inserm, Radiothérapie Moléculaire et Innovation Thérapeutique
Exploring Deep Registration Latent Spaces
Jul 23, 2021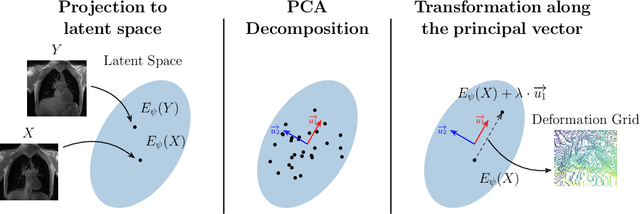
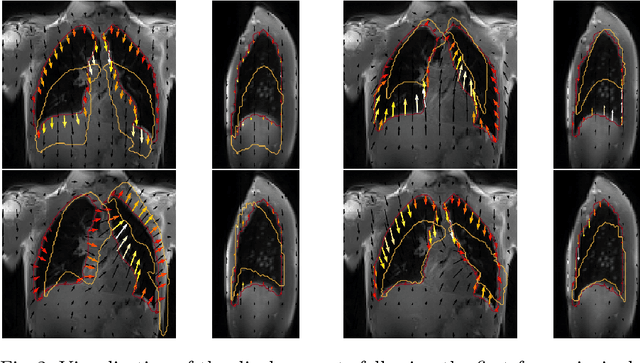
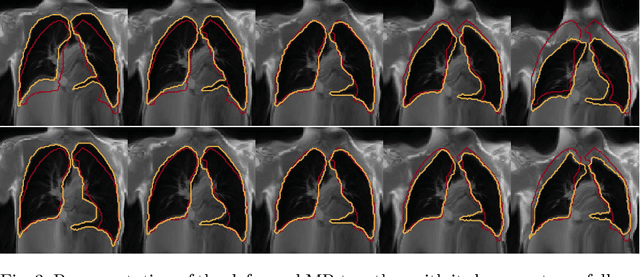
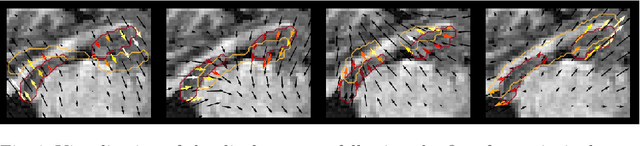
Abstract:Explainability of deep neural networks is one of the most challenging and interesting problems in the field. In this study, we investigate the topic focusing on the interpretability of deep learning-based registration methods. In particular, with the appropriate model architecture and using a simple linear projection, we decompose the encoding space, generating a new basis, and we empirically show that this basis captures various decomposed anatomically aware geometrical transformations. We perform experiments using two different datasets focusing on lungs and hippocampus MRI. We show that such an approach can decompose the highly convoluted latent spaces of registration pipelines in an orthogonal space with several interesting properties. We hope that this work could shed some light on a better understanding of deep learning-based registration methods.
Weakly supervised pan-cancer segmentation tool
May 10, 2021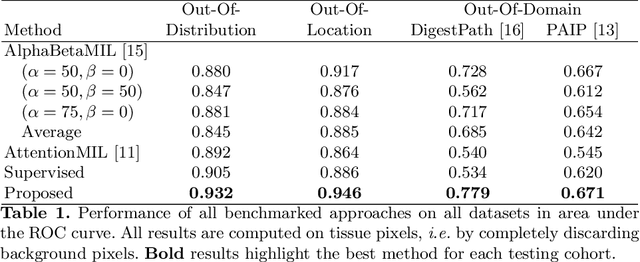
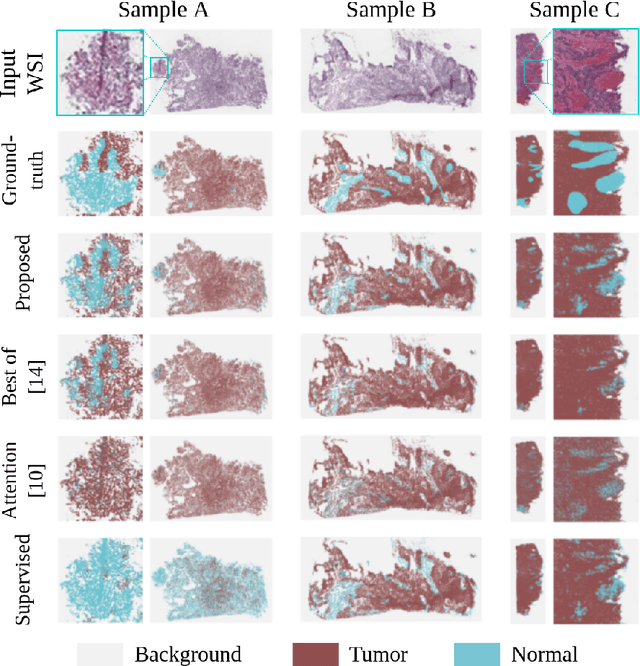
Abstract:The vast majority of semantic segmentation approaches rely on pixel-level annotations that are tedious and time consuming to obtain and suffer from significant inter and intra-expert variability. To address these issues, recent approaches have leveraged categorical annotations at the slide-level, that in general suffer from robustness and generalization. In this paper, we propose a novel weakly supervised multi-instance learning approach that deciphers quantitative slide-level annotations which are fast to obtain and regularly present in clinical routine. The extreme potentials of the proposed approach are demonstrated for tumor segmentation of solid cancer subtypes. The proposed approach achieves superior performance in out-of-distribution, out-of-location, and out-of-domain testing sets.
Deep learning based registration using spatial gradients and noisy segmentation labels
Oct 21, 2020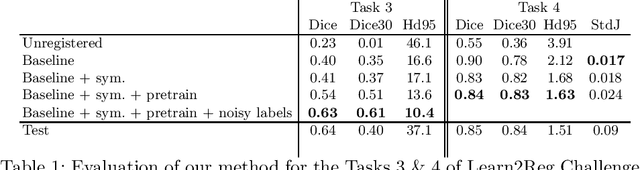
Abstract:Image registration is one of the most challenging problems in medical image analysis. In the recent years, deep learning based approaches became quite popular, providing fast and performing registration strategies. In this short paper, we summarise our work presented on Learn2Reg challenge 2020. The main contributions of our work rely on (i) a symmetric formulation, predicting the transformations from source to target and from target to source simultaneously, enforcing the trained representations to be similar and (ii) integration of variety of publicly available datasets used both for pretraining and for augmenting segmentation labels. Our method reports a mean dice of $0.64$ for task 3 and $0.85$ for task 4 on the test sets, taking third place on the challenge. Our code and models are publicly available at https://github.com/TheoEst/abdominal_registration and \https://github.com/TheoEst/hippocampus_registration.
Weakly supervised multiple instance learning histopathological tumor segmentation
Apr 21, 2020
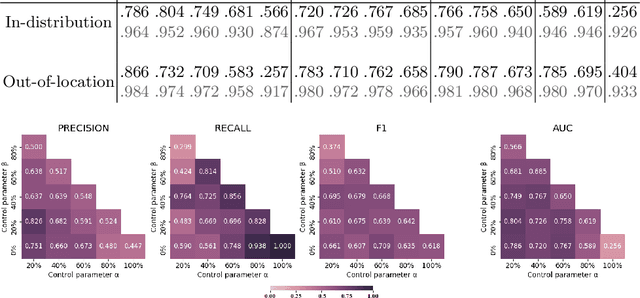
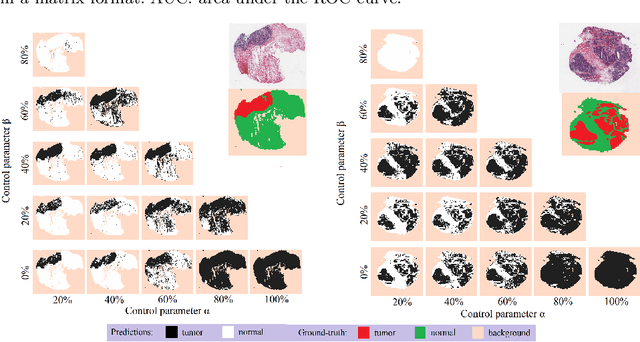
Abstract:Histopathological image segmentation is a challenging and important topic in medical imaging with tremendous potential impact in clinical practice. State of the art methods relying on hand-crafted annotations that reduce the scope of the solutions since digital histology suffers from standardization and samples differ significantly between cancer phenotypes. To this end, in this paper, we propose a weakly supervised framework relying on weak standard clinical practice annotations, available in most medical centers. In particular, we exploit a multiple instance learning scheme providing a label for each instance, establishing a detailed segmentation of whole slide images. The potential of the framework is assessed with multi-centric data experiments using The Cancer Genome Atlas repository and the publicly available PatchCamelyon dataset. Promising results when compared with experts' annotations demonstrate the potentials of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge